Abstract
Trichiurus japonicus is one of Trichiuridae and widely distributed in the North Pacific Ocean. In this study, we described the complete mitochondrial genome of Trichiurus japonicus. The genome is 16797 bp in length, encoding the standard set of 13 protein-coding genes, 21 transfer RNA genes, two ribosomal RNA genes, and a non-coding D-loop, with circular organization. The overall base composition of the whole mitochondrial genome was A (28.7%), T (25.83%), G (16.2%), and C (29.27%) with an AT bias of 54.53%. The longest protein-coding gene of these species was ND5, whereas the shortest ATP8.
Cutlassfishes (Trichiuridae) are bentho-pelagic predators, inhabiting continental shelves and slopes of the world (Nelson Citation2006). They are a diverse group of demersal fishes including more than 30 species in nine genera (Nakamura and Parin Citation1993). Among the known genera, the Trichiurus genus with at least 10 species accounts for a significant proportion of commercial catches in world fisheries production. In the western North Pacific, Trichiurus japonicus is included in T. lepturus and is arguably treated as a junior synonym (Tucker Citation1956; Nelson Citation2006). In fact, T. japonicus from Japan and the East China Sea were reported to have higher numbers of dorsal fin rays and vertebrae. Hence, some taxonomists proposed that T. japonicus and T. lepturus are two valid species based on distinguishable morphological characters (Lee et al. Citation1977). Here, we sequenced and annotate mitogenome of T. japonicus form South China Sea to provide molecular information for genetically understanding of cutlassfishes fish.
The specimens of T. japonicus were collected from the South China Sea (20°30′N, 118°30′E) in 15th September, 2017. Whole genomic DNA was extracted from muscle tissue of one specimen of T. japonicus using TIANamp Marine Animals DNA Kit (TIANGEN, China). The concentration for use as a PCR template was adjusted to an A260 of about 0.05–0.2. The collected specimen and extracted DNA were stored in Guangdong Provincial Key Laboratory of Fishery Ecology and Environment. The complete mitochondrial genomes of T. japonicus was sequenced using PCR primers designed from highly conserved regions of transfer RNA (tRNA) sequences of related species (Liu and Cui Citation2009) with additional specific primers designed as required from sequences already obtained. Long-PCR amplifications were performed by thermo-cycling using five pairs of primers and PCR amplicons were subjected to build up genomic library and pair-end sequencing using MiSeq. The COI sequence of T. japonicus was used as reference seeds for iterative assembly using MITObim v.1.8 (Hahn et al. Citation2013). SeqMan v.7.1.0 was used for the mitogenome assembly and annotation (Swindell and Plasterer Citation1997). Transfer RNA genes were predicted using online software tRNAScan-SE 1.21 (Lowe and Eddy Citation1997). All Protein coding gene (PCGs) are aligned independently, then concatenated to be applied for phylogenetic reconstruction with other Scombriformes in MrBayes v 3.12 (Ronquist and Huelsenbeck Citation2003) using relaxed clock model.
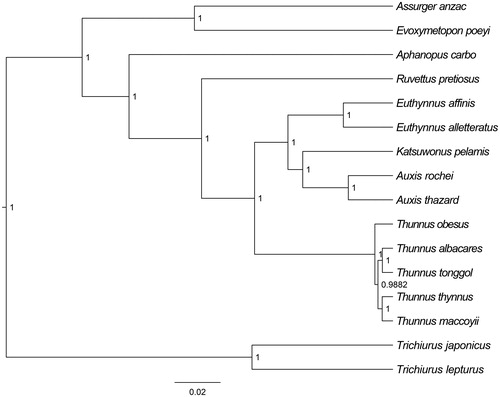
The T. japonicus mitochondrial genome forms a 16797 bp closed loop (GenBank accession number MK292708). The overall base composition of the whole mitochondrial genome was A (28.7%), T (25.83%), G (16.2%), and C (29.27%) with an AT bias of 54.53%. This mitochondrial genome represents a typical Trichiurus mitochondrial genome and matches with the T. lepturus genome (Liu et al. Citation2013), in which it comprises 13 protein-coding genes, 21 transfer RNA genes, and two ribosomal RNA genes (12S rRNA and 16S rRNA) and one A + T-rich region which could also be termed as control region. The ATG initiation codon are used in all protein-coding genes except COX1 (GTG), ND1 (ATA), ND3 (ATT), and the stop codons of all the 13 protein-coding genes were complete. Meanwhile, the longest protein-coding gene of these species was ND5 (1746 bp), whereas the shortest ATP8 (168 bp). lrRNA and srRNA genes are 1703 bp and 958 bp in length separately, and the length of D-loop is 508 bp. All the 21 typical tRNAs possess a complete clover leaf secondary structure, ranging from 66 bp to 74 bp. The Bayesian inference phylogenetic tree showed that T. japonicus firstly grouped with species of T. lepturus (). We have the confidence to construct phylogenetic trees, based on the complete the mitochondrial genomes, but the evolution history of cutlassfish still needs future research to be clearly resolved.
Figure 1. The Bayesian inference phylogenetic tree for Scombriformes based on mitochondrial PCGs and rRNAs concatenated dataset. The gene’s accession numbers for tree construction are listed as follows: Evoxymetopon poeyi (AP012509), Assurger anzac (AP012508), Aphanopus carbo (AP012944), Thunnus thynnus (KF906720), Thunnus obesus (JN086152), Thunnus maccoyii (KF925362), Thunnus tonggol (HQ425780), Thunnus albacares (GU256528), Euthynnus affinis (AP012946), Katsuwonus pelamis (KM605252), Ruvettus pretiosus (AP012506), Euthynnus alletteratus (AB099716), Auxis rochei (AB103468), Auxis thazard (KP259551), Trichiurus lepturus (JX477078).
Disclosure statement
None of the co-authors has any conflict of interest to declare. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Lee SC, Chang KH, Wu WL, Yang HC. 1977. Formosan ribbonfishes (Perciformes: Trichiuridae). Bull Inst Zool Acad Sin. 16:77–84.
- Liu Y, Cui Z. 2009. The complete mitochondrial genome sequence of the cutlassfish Trichiurus japonicus (Perciformes: Trichiuridae): genome characterization and phylogenetic considerations. Mar Genomics. 2(2):133–142.
- Liu X, Guo Y, Wang Z, Liu C. 2013. The complete mitochondrial genome sequence of Trichiurus nanhaiensis (Perciformes: Trichiuridae). Mitochondrial DNA. 24(5):516–517.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Nakamura I, Parin NV.1993.FAO species catalogue, Snake mackerels and cutlassfish of the world (Families Gempylidae and Trichiuridae). FAO Fish. Synos. 125:61–107.
- Nelson JS. 2006. Fishes of the world. 4th ed. New York: John Wiley and Sons Inc.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Swindell SR, Plasterer TN. 1997. Seqman, contig assembly. In: Swindell SR, editor. Sequence data analysis guidebook. Totowa (NJ): Springer; p. 75–89.
- Tucker, D. W., 1956. Studies on the trichiuroid fishes a preliminary revision of the family Trichiuridae. Bull Br Mus Zool. 4:73–103.
