Abstract
Populus pamirica Kom. was merely found in Pamir and formed a rare type of riparian forest. Here we assembled the complete chloroplast genome of P. pamirica, which is 157,418 bp in length, with 129 functional genes including eight rRNA, 36 tRNA, and 85 protein-coding genes. The GC content of P. pamirica is 36.6%. Our phylogenetic inference suggested that P. pamirica had close relationship with P. szechuanica. Further studies such as combining nrDNA data are needed to confirm the systematic position of P. pamirica.
Populus L. is a woody genus, which was classified as six sections. Populus pamirica Kom. belonging Sect. Tacamahaca is similar to P. pilosa and P. laurifolia in morphology. P. pamirica is found in Pamir Plateau, including southern Xinjiang, Pakistan and Tajikistan. This species grows along the river and on the forest edge of spruce. Within Sect. Tacamahaca, ten complete chloroplast genomes have been reported and nine of which are from America and East Asia, only one in Central Asia (Tuskan et al. Citation2006; Zhang et al. Citation2018). Thus, the phylogenetic relationship of Central Asian species still remains poorly resolved. In this study, we report the complete chloroplast genome sequence of P. pamirica.
Fresh leaves were collected from natural individual of P. pamirica in Khunjerab National Park of Pakistan (N 36°47′31″, E 74°53′74″), and the voucher specimens were deposited at Herbarium, Institute of Botany, CAS (PE) (collection number is FPH-0368). CTAB method (Doyle and Doyle Citation1987) was used to extract the total genome DNA which was then send to Novogene Company (http://www.novogene.com, China) for 2 × 150 bp pair end sequencing using Illumina HiSeq 4000 platform. Referring to the genome of P. trichocarpa, the Map tool of Geneious R11 (Kearse et al. Citation2012) was used to filter available reads, which were used for de novo assembly using Geneious R11, then we used Fine Tuning function to bridge gaps. Plann (Huang and Cronk Citation2015) was employed to annotate the chloroplast genome sequence, and the verification was also performed using Genious R11.
The complete chloroplast genome of P. pamirica is 157,418 bp in length and contains a large single-copy (LSC) region of 85,401 bp, a small single-copy (SSC) region of 16,717 bp, and a pair of inverted repeat (IR) regions of 27,650 bp. This plastome contains 129 genes, including 85 protein-coding genes, eight ribosomal RNA genes and 36 transfer RNA genes. The GC content of the total sequence is 36.6%. The IR regions (41.9%) possess higher GC content than the LSC (34.3%) and SSC (30.5%) regions.
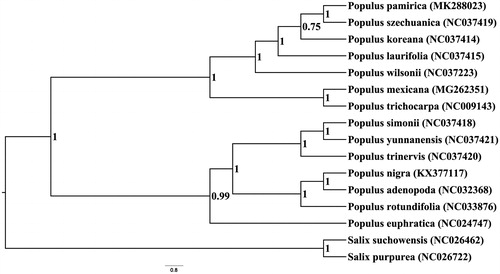
To infer the phylogenetic position of P. pamirica, we constructed Bayesian inference using Mrbayes 3.2.6 (Ronquist and Huelsenbeck Citation2003) based on 16 published complete chloroplast genome sequences of Populus and Salix. The sequences were aligned by MAFFT v6.833 (Katoh et al. Citation2005). Our result firstly showed the phylogenetic position of P. pamirica, which fell into a clade formed by P. szechuanica, P. laurifolia and P. koarna along with this species
Figure 1. Bayesian phylogram of Populus pamirica as well as other related species inferred from the complete plastome sequences.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Eckenwalder JE. 1984. Natural intersectional hybridization between North American species of Populus (Salicaceae) in sections Aigeiros and Tacamahaca. II. Taxonomy. Can J Bot. 62:325–335.
- Feng JJ, Jiang DC, Shang HY, Dong M, Wang GN, He XY, Zhao CM, Mao KS. 2013. Barcoding Poplars (Populus L.) from Western China. PLoS One. 8:e71710.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 313:1596–1604.
- Wang ZS, Du SH, Dayanandan S, Wang DS, Zeng YF, Zhang JG. 2014. Phylogeny reconstruction and hybrid analysis of Populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PLoS One. 9:e103645.
- Zhang L, Xi ZX, Wang MC, Guo XY, Ma T. 2018. Plastome phylogeny and lineage diversification of Salicaceae with focus on poplars and willows. Ecol Evol. 0:1–8.
