Abstract
Musa banksii is a species of wild banana in the genus Musa, native to New Guinea, Manus, northern Australia and some islands of eastern Indonesia. In this study, the chloroplast genome sequence of M. banksii been assembled and characterized by Illumina sequencing. The complete chloroplast genome was 170,010 bp in length, containing a large single copy region (LSC) of 88,373 bp and a small single copy region (SSC) of 10,769 bp, which were separated by a pair of 35,434 bp inverted repeat regions (IRs). A total of 136 genes were predicted, including 113 unique genes consisting of 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Maximum-likelihood phylogenetic analysis of 11 representative plastomes within the order Zingiberales suggests that M. banksii closely related to the Musa ornata and Musa acuminata.
Musa banksii is also called maroon-stem banana, it is a species of wild banana in the genus Musa (Argent 1976), sometimes treated as a subspecies Musa acuminata spp. banksii, native to New Guinea, Manus, northern Australia and some islands of eastern Indonesia (Denham and Donohue Citation2009). Musa banksii was considered to be the original parent of cultivar bananas, it exhibits slightly waxy leaf, predominantly brown-blackish pseudostems, large bunches with splayed fruits, and non-imbricated yellow bracts (Rouard et al. Citation2018), it is slightly smaller from commercially grown species and is starchy and packed with seed fruits which grow its length from 8 to 14 cm (David Citation2007), it could be valuable for banana breeding (Jarret et al. Citation1992). In this study, we characterized the complete chloroplast genome sequence of M. banksii as a resource for future genetic studies on related species
Leaf samples from accessions M. banksii were supplied by the CRB-Plantes Tropicales Antilles CIRAD field collection based in Guadeloupe. Genomic DNA was extracted from dry leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina technology, a library with the insertion size of 800 bp was constructed, and high-throughput DNA sequencing was performed on an Illumina Hiseq 2000 platform, generating approximately 6 Gb of sequence data. Illumina sequencing data were assembled using CLC Genomic Workbench v3.6 (http://www.clcbio.com). The chloroplast genome was annotated using DOGMA (Wyman et al. Citation2004) with manual correction. The accurate new annotated complete chloroplast genome was submitted to GenBank with accession number MK210631.
The complete chloroplast genome sequence of M. banksii was 170,010 bp in length, with a large single-copy (LSC) region of 88,373 bp, a small single-copy (SSC) region of 10,769 bp, and two inverted repeat (IRs) regions of 35,434 bp each. A total of 136 genes were predicted, including 113 unique genes consisting of 79 protein-coding genes, 4 ribosomal RNA (rRNA) genes and 30 transfer RNA (tRNA) genes. The base composition of the chloroplast genome is A (31.43%), G (18.16%), C (18.65%), and T (31.76%), the overall GC content was 36.81%.
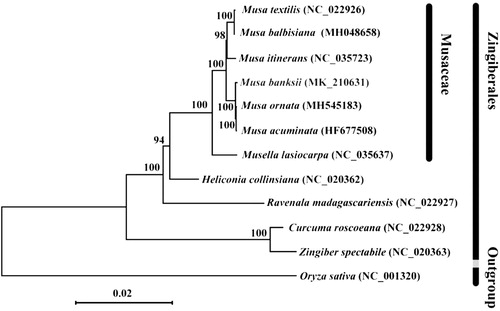
To understand the phylogenetic position of M. banksii within the order Zingiberales, chloroplast genome sequences of ten other representative species in Zingiberales (Musa banksii, Musa ornata, Musa balbisiana, Musa textilis, Musa itinerans, Musa acuminata, Musella lasiocarpa, Heliconia collinsiana, Curcuma roscoeana, Ravenala madagascariensis, Zingiber spectabile), and Oryza sativa as the outgroup (). A total of 12 selected complete chloroplast genome sequences was aligned using MAFFT (Katoh and Standley Citation2013). Maximum-likelihood (ML) analysis was performed using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates. Our results showed that M. banksii clustered within Musa ornata and Musa acuminata with strong bootstrap support (). The phylogenetic position of other species consists with previous study (Niu et al. Citation2018).
Figure 1. Phylogenetic relationships of 12 species based on chloroplast genome sequences. The taxon in red colour is the new chloroplast genome reported in this study. Bootstrap support is indicated for each branch. GenBank accession number: Musa banksii (MK210631), Musa ornata (MH545183), Musa balbisiana (MH048658), Musa textilis (NC_022926), Musa itinerans (NC_035723), Musa acuminata (HF677508), Musella lasiocarpa (NC_035637), Heliconia collinsiana (NC_020362), Curcuma roscoeana (NC_022928), Ravenala madagascariensis (NC_022927), Zingiber spectabile (NC_020363), Oryza sativa (NC_001320).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Argent GCC. 1976. The wild bananas of Papua New Guinea. Notes Roy Bot Gard Edinburgh. 35:77–114.
- David H. 2007. Fruits of the forest. Australian Geographic. 86:20–21.
- Denham T, Donohue M. 2009. Pre-Austronesian dispersal of banana cultivars west from new guinea: linguistic relics from eastern Indonesia. Archaeol Oceania. 44:18–28.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quan- tities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jarret RL, Gawel N, Whittemore A, Sharrock S. 1992. RFLP-based phylogeny of Musa species in Papua New Guinea. Theor Appl Genet. 84:579–584.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Niu YF, Gao CW, Liu J. 2018. The complete chloroplast genome sequence of wild banana, Musa balbisiana variety ’Pisang Klutuk Wulung’ (Musaceae). Mitochondrial DNA B. 3:460–461.
- Rouard M, Droc G, Martin G, Sardos J, Hueber Y, Guignon V, Cenci A, Geigle B, Hibbins MS, Yahiaoui N, et al. 2018. Three new genome assemblies support a rapid radiation in Musa acuminata (Wild Banana)). Gen Biol Evol. 10:3129–3140.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
