Abstract
The study determined the first complete mitochondrial genome (mitogenome) of the polychaete, Marphysa tamurai. A total of 21,346 reads were generated by Illumina HiSeq2500 platform with an average depth of 175.97×. The mitogenome of M. tamurai was 15,163 bp in size and consists of 38 typical genes, including 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes and 1 D-loop region. All the 38 genes were encoded on the heavy strand whose nucleotide compositions were 30.23% of A, 30.40% of T, 12.59% of G, 26.78% of C, showing a lower content of G + C (39.37%). Phylogenetic analysis showed that M. tamurai had a closer relationship with Marphysa sanguinea of Nereididae.
Marphysa is one of the nine genera in the family Eunicidae of Polychaeta (Zanol et al. Citation2014). It comprises about 60 species in the genus Marphysa and is widely used for bait known as blood worms by recreational fishermens (Saito et al. Citation2014; Zanol et al. Citation2014; Liu et al. Citation2017). Most of the Marphysa species are large-sized with high economic value and are traded as a commodity (Li et al. Citation2016). Marphysa species is distributed worldwide predominantly in estuarine or sheltered habitats, and plays important roles in seabed or intertidal communities (Zanol et al. Citation2016). In China seas, it occurs regularly in coastal and estuarine tidal flats, especially along the southeast coast. Polychaeta systematics, molecular phylogeney, and ingroup relationships are a matter of ongoing debates in recent analyses (Sekar et al. Citation2016). The complete mitochondrial genome can provide more valuable information for those studies (Chen et al. Citation2016). Here, we determined the first complete mitogenome of Marphysa tamurai and attempted to provide more information for studies on the systematics, phylogeny, and ingroup relationships within the family Eunicidae.
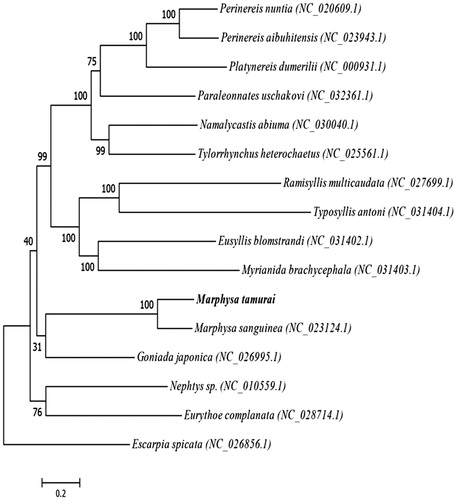
The specimen was collected from shallow sandbeach near Hailing Bay in southern China sea, the sampling spot (21°61′N, 111°79′E) was used with the coordination record of the GPS. The specimen was preserved in 95% ethanol and stored at −70 °C in our laboratory (The Key Laboratory for Marine Estuary Fishery Resources Protection of Yangjiang, Yangjiang city, Guangdong province, China). The total genomic DNA was extracted from muscle tissue using the salting-out procedure (Howe et al. Citation1997). A total of 21,346 reads were generated by Illumina HiSeq2500 platform with an average depth of 175.97×. The complete mitogenome of M. tamurai (Genbank accession number NC_037236) is 15,163 bp in length, including the typical structure of 2 ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA), 13 protein-coding genes (PCGs), and 1 D-loop region. All 38 genes are encoded on the heavy strand whose nucleotide composition is 30.23% A, 26.78% C, 12.59% G, and 30.40% T, showing a higher content of A + T (60.63%) than G + C (39.37%). There are a total of 13 bp gene overlapping regions at 5 locations and 202 bp intergenic spacers at 15 locations. Among 13 protein-coding genes, 6 share the complete termination codon TAA and 3 with TAG, while others terminate with an incomplete termination codon T or TA, all these 13 genes use ATG as an initiation codon. Two rRNA genes are located between the tRNA-Met and tRNA-Leu genes, and separated by the tRNA-Val gene. The 22 tRNA genes vary from 59 to 69 bp in length. The phylogenetic relationships of M. tamurai were inferred by its 13 protein-coding genes compared with other 14 complete or partial mitogenomes (downloaded from NCBI Genbank database) by Maximum-Likelihood (ML) methods in RAxML8.1.5 (Stamatakis Citation2006, Citation2014), and the Bootstrap value was 1000. Phylogenetic analysis showed that M. tamurai had a closer relationship with Marphysa sanguinea of Nereididae ().
Figure 1. Maximum-Likelihood (ML) phylogenetic tree of M. tamurai with other 14 polychaeta species, GenBank accession numbers are listed parenthetically. This tree was determined by RAxML8.1.5. Numbers associated with branches are ML bootstrap support values based on 1000 replicates. Escarpia spicata is set as outgroup. The scale bar represent 0.2, which is an indicator of genetic distance based on branch length.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Chen XH, Li MM, Liu HP, Li B, Guo L, Meng ZN, Lin HR. 2016. Mitochondrial genome of the polychaete Tylorrhynchus heterochaetus (Phyllodocida, Nereididae). Mitochondrial DNA Part A. 27:3372–3373.
- Howe JR, Klimstra DS, Cordon-Cardo C. 1997. DNA extraction from paraffin-embedded tissues using a salting-out procedure: a reliable method for PCR amplification of archival material. Histol Histopathol. 12:595–601.
- Li S, Chen Y, Zhang M, Bao X, Li Y, Teng W, Liu Z, Fu C, Wang Q, Liu W. 2016. Complete mitochondrial genome of the marine polychaete, Marphysa sanguinea (Polychaeta, Eunicida). Mitochondrial DNA Part A. 27:42–43.
- Liu YB, Hutchings P, Sun SC. 2017. Three new species of Marphysa Quatrefages, 1865 (Polychaeta: Eunicida: Eunicidae) from the south coast of China and redescription of Marphysa sinensis Monro, 1934. Zootaxa. 4263:228–250.
- Saito H., Kawai K., Umino T., Imabayashi H. 2014. Fishing bait worm supplies in Japan in relation to their physiological traits. Mem Museum Victoria. 71:279–287.
- Sekar V, Rajasekaran R, Prasannakumar C, Sankar R, Sridhar R, Sachithanandam V. 2016. Morphological and COI sequence based characterisation of marine polychaete species from Great Nicobar Island, India. In: Trivedi S, Ansari A, Ghosh S, Rehman H. (eds) DNA barcoding in marine perspectives. Cham: Springer; p. 89–111.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zanol J, Halanych KM, Fauchald K. 2014. Reconciling taxonomy and phylogeny in the bristleworm family Eunicidae (polychaete, Annelida). Zool Scripta. 43:79–100.
- Zanol J, Silva TDS, Hutchings P. 2016. Marphysa (Eunicidae, polychaete, Annelida) species of the Sanguinea group from Australia, with comments on pseudo-cryptic species. Invertebr Biol. 135:328–344.
