Abstract
Backcross sea bream Acanthopagrus schlegelii (female) and [A. schlegelii (female) and P. major (male)] (male) have the desirable trait of heat tolerance and growth performance. In this study, the complete mitochondrial genome of the backcross sea bream was obtained by the next-generation sequencing. The genome is 16,649 bp in length, including 2 ribosomal RNA genes, 13 proteins-coding genes, 22 transfer RNA genes, and a non-coding control region. Sequence analysis showed that the overall base composition is 28.0% for A, 27.9% for T, 16.2% for C, and 27.9% for G.
Keywords:
Acanthopagrus schlegelii and Pagrus major, both belong to Sparidae in the order Perciformes, which are widely distributed in the West Pacific coast from Japan, Korea to the East China Sea. As the hybrid of A. schlegelii (♀) × P. major (. has faster growth, backcross sea bream A. schlegelii (female) and [A. schlegelii (female) and P. major (male)] (male) has heat tolerance and growth performance. There is no report of the complete genome of this hybrid. Therefore, the mitochondrial genome of this species is important, and this mitochondrial genome can be utilized for taxonomic evolution, population genetic analysis, etc. (Liu et al. Citation2016).
The specimen collected from Jiangsu Province seawater aquaculture technology and seedling center (Nantong, China). Its latitude and longitude are N32°04′35.75″, E121°36′23.25″. The specimen’s tail fin is stored in Jiangsu Key Laboratory for Genetics and Breeding of Marine Fishes (Nantong, China). Its storage code is APAbackcross2016. The genome is 16,649 bp in length (Genbank, MK112875), including 2 ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and a non-coding control region. Sequence analysis showed that the overall base composition is 28.0% for A, 27.9% for T, 16.2% for C, and 27.9% for G, respectively. Twelve PCGs, 14 tRNA genes and two rRNA genes were located on the heavy strand (H-strand), while one PCG (ND6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, tRNAPro) on the light strand (L strand). Five PCGs (ND1, ND2, COIII, ND4L, ND5, and ND6) were terminated with TAA stop codon, ATP8 ended with TAG. On the other hand, remaining four PCGs (ATP6, ND3, ND4, and CYTB) ended with the incomplete stop codon represented as a single T. COI was terminated AGG. COII was terminated AGA. Frame overlapping took place in three pairs of PCGs. ATP8 and ATP6 overlapped by 7 nucleotides (nt), ND4L and ND4 for 7 nt, and ND5 and ND6 (encoded on opposing stand) with 4 nt.
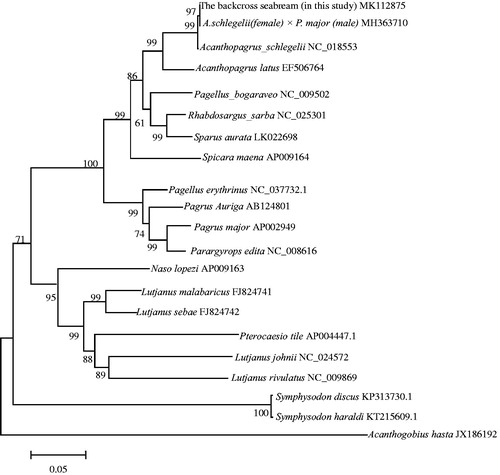
The determination of the taxonomic status of backcross sea bream was conducted by using mitochondrial genomic data sets and 20 other fish species from the GeneBank database. Three primary branches were supported (). Among them, backcross sea bream in this study, A. schlegelii, A. schlegelii ♀ × P. major ♂. A. latus, Pagellus bogaraveo, Rhabdosargus sarba, Sparus aurata, Spicara maena, P. auriga, P. erythrinus, P. major, Parargyrops edita, clustered into one branch, with all eleven taxa belonging to Sparidae. The backcross sea bream A. schlegelii (female) and [A. schlegelii (female) and P. major (male)] (male) has a closer relationship with A. schlegelii ♀ × P. major ♂ (Zhu et al. Citation2018) (female parent) than A. schlegelii (male parent), which means that backcross in Sparidae is also by maternal inheritance characteristics of the mitochondria. Pterocaesio tile, Lutjanus johnii, L. rivulatus, L. malabaricus, L. sebae clustered into another branch, with all the five taxa belonging to Lutjanidae. L. johnii, L. rivulatus, L. malabaricus, L. sebae clustered into another branch, with all four taxa belonging to Lutjanidae. In addition to the above, Symphysodon haraldi, S. discus, and A. hasta were the outgroup.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Liu Y, Hou J, Wang G, Zhang X, Liu H. 2016. A species-specific primer pair for distinguishing between Paramisgurnus dabryanus and Misgurnus anguillicaudatus based on mitochondrial DNA polymorphisms. Mitochondrial DNA Part A. 27:2326.
- Zhu F, Xue L, Jia C, Zhang Z, Chen S, Zhang Z, Meng Q, Sun R. 2018. The complete mitochondrial genome of the hybrid of Acanthopagrus schlegelii (♀) × Pagrus major (♂) with phylogenetic analysis. Mitochondrial DNA Part B. 3:1067–1068.