Abstract
The mitochondrial genomes of Seriola quinqueradiata and S. lalandi were 16,539 and 16,535 base pairs long, respectively, whose organizations were similar to those of other bony fishes. The sequence similarity of two species was 93% in the mitochondrial whole genome. Between each coding gene, rRNA, and control region, the highest similarity was 98% in ATP8 and lowest similarity was 87% in the control region. Phylogenetic analysis of Carangidae mt 12 protein-coding genes showed that Seriola and Carangoides do not form a monophyletic genus.
Seriola is a genus of Carangidae in Perciforms, known as amberjacks. Japanese and Yellowtail amberjack, Seriola quinqueradiata and Seriola lalandi, respectively, are morphologically very similar, produced through aquaculture, and valuable foods in Korea and Japan.
For complete mitochondrial (mt) genome analysis, we extracted genomic DNA from blood samples of S. quinqueradiata and S. lalandi provided by the Jeju Fisheries Research Institute. Genomic DNA was stored at −80 °C.
The mt genomes of S. quinqueradiata and S. lalandi were 16,539 and 16,535 base pairs, respectively. The mt genome sequences were deposited in the GenBank database (Accession No. MK158067 and MK158068, respectively). Like the mt genomes of other bony fishes, both genomes contain 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, and a non-coding control region.
The overall base compositions were as follows: S. quinqueradiata: A, 26.54%; C, 30.26%; G, 18.08%; T, 25.13%; S. lalandi: A, 26.57%; C, 30.22%; G, 17.91%; T, 25.30%. The respective proportions of A and T in the base composition were relatively low. The respective proportions of C and G were slightly higher than in S. dumerili (AB517558) and S. rivoliana (KP733847).
The PCGs of S. quinqueradiata and S. lalandi begin with ATG except for cytochrome c oxidase subunit 1, which begins with GTG and ends with TAG, TAA, or incomplete termination codon, T. Incomplete termination codons, which are added through posttranscriptional polyadenylation (Ojala et al. Citation1981), are common in bony fish mt genomes (Ishiguro et al. Citation2001; Miya et al. Citation2003; Oh et al. Citation2008).
The mt genomes of S. quinqueradiata and S. lalandi contain 22 tRNA genes. We identified 2 types of trnL(UUR and CUN) and trnS(UCN and AGY) and 3 clusters (IQM, WANCY, and HSL), which are conserved in Seriola fishes and bony fish mt genomes (Ishiguro et al. Citation2001; Miya et al. Citation2003; Oh and Jung Citation2008).
Examination of the differences in mt genes and the control region revealed that the 13 PCGs ranged between 87% (control region) and 98% (ATPase subunit 8 and 12S rRNA) identity. Overall, the mt genome of S. quinqueradiata and S. lalandi showed 93% identity.
To phylogenetically analyze Carangidae fishes, we constructed a maximum likelihood tree using MEGA X software (Kumar et al. Citation2018) with 12 PCGs excluding NADH dehydrogenase subunit 6 gene because of its heterogeneous base composition and consistently poor phylogenetic performance (Zardoya and Meyer Citation1996; Miya and Nishida Citation2000). GTR + G + I mode was selected as the best substitution model based on AIC criterion (Akaike Citation1973).
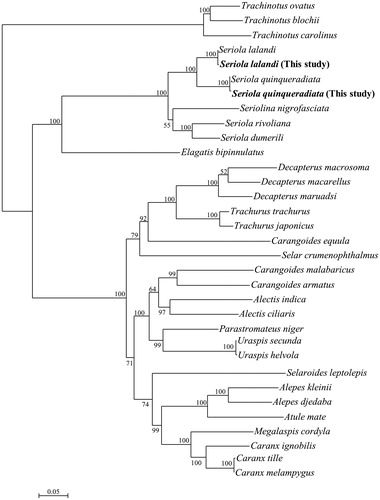
Phylogenetic analysis of Carangidae showed that Seriola is not a monophyletic group (). The black-banded trevally, Seriolina nigrofasciata, was between S. dumerili + S. rivoliana and S. quinqueradiata + S. lalandi. This result slightly differed from that of a previous study (Santini and Carnevale Citation2015) showing that S. nigrofasciata was the first lineage to branch off and Seriola formed a diverse clade with Naucrated doctor. Carangoides was also a non-monophyletic genus in this study and according to Santini and Carnevale (Citation2015). Our phylogenetic analysis suggested that Carangidae fish phylogeny should be re-examined.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Akaike H. 1973. Information theory as an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second international symposium on information theory. Budapest: Akademiai Kiado.
- Ishiguro N, Miya M, Nishida M. 2001. Complete mitochondrial DNA sequence of ayu Pleoglossus altivelis. Fisheries Sci. 67:474–481.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Miya M, Nishida M. 2000. Use of mitogenome information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogent Evol. 17:437–455.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138.
- Oh DJ, Jung YH. 2008. The mitochondrial genome of the threespot wrasse Halichoeres trimaculatus (Perciformes, Labridae). Genes Genom. 30:113–120.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Santini F, Carnevale G. 2015. First multilocus and densely sampled timetree of trevallies, pompanos and allies (Carangoidei, Percomorpha) suggests a Cretaceous origin and Eocene radiation of a major clade of piscivores. Mol Phylogenet Evol. 83:33–39.
- Zardoya R, Meyer A. 1996. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol Biol Evol. 13:933–942.