Abstract
Monilinia fructicola is a plant pathogenic fungus usually causing brown rot in the stone fruit crops. Here, the complete mitochondrial genome of M. fructicola is first reported, and the length of the circular genome was 163,790 bp and the content of GC was 31%. Meanwhile, it contained 28 transfer RNA (tRNA) genes, 30 protein-coding genes (PCGs) with many intron sequences and 2 ribosomal RNA (rRNA) genes. Phylogenetic analysis with 15 Leotiomycetes species indicated that M. fructicola was clustered in the Sclerotiniaceae family and was closely narrowed to Botryotinia fuckeliana and Sclerotinia scleroticrum. In addition, the valuable gene information of the mitochondrial genome was provided, and the study on the control of this destructive pathogen would be facilitated.
Brown rot is one of the most important reasons for economic loss in the stone fruit crops in many countries, such as Korea, Australia and China (Hu et al. Citation2011; Choi et al. Citation2016; Tran et al. Citation2017), and it is mainly caused by Monilinia spp. that usually attacks plants belonging to Rosaceae, Ericaceae and Empetraceae (Oliveira et al. Citation2016). Among the genus Monilinia, M. fructicola was considered as the most highly infectious pathogen of the stone fruit crops, and it was found that this pathogen infected these plants at different stages, including bloom, twig, and fruit (Martini and Mari Citation2014). Compared with Monilinia laxa and Monilinia fructigena, M. fructicola was the most hard to control because of its sexual recombination and anastomosis, and at the same time, it had a greater genetic change and potential to overcome genetic barriers (Grzegorczyk et al. Citation2017; Tran et al. Citation2017). Therefore, this fungus was listed in the European and Mediterranean Plant Protection Organization (EPPO) A2 list of quarantine pests in Europe (EFSA Panel on Plant Health (PLH) Citation2011). Additionally, M. fructicola was an ideal material to study the fungus disease control of plants. Here, the complete mitochondrial genome of M. fructicola was recovered by Illumina sequencing data, and its genome profile and phylogenetic evolution were analyzed.
The sample of M. fructicola adopted in this study was isolated from Yunnan,China (25.05°N, 102.72°W) and obtained from the infected stone fruit by Doctor Yuanbing Wang, and axenic culture was transplanted to Potato Dextrose Agar (PDA) slants and stored at 4 °C. Then, the total genomic DNA of M. fructicola was extracted by MiniBEST Plant Genomic DNA Extraction Kit (TaKaRa, China) according to the manufacturer’s instructions and deposited in Research Center of Cordyceps Development and Utilization of Kunming. Later, agarose gel electrophoresis was applied to verify the quality and integrity of its DNA, and the purified DNA was adopted to be built in a 270 bp library and sequenced with Illumina HiSeq2500 platform by Biomarker Co. Ltd (Beijing, China). Based on the high-quality reads, sequences were assembled by SPAdes 3.9.0 with default parameter (Bankevich et al. Citation2012). The mitochondrial genome was annotated using MFannot tool and AEWEN Web server with manual correction.
The complete mitogenome of M. fructicola was first sequenced and annotated with GenBank accession number of MK163638. The mitochondrial genome of M. fructicola was 163,790 bp in length, with a large number of repeated sequences and introns. It contained 2 ribosomal RNA (rRNA), 28 transfer RNA (tRNA), and 30 protein-coding genes (PCGs) which consisted of 7 NAD genes, 1 COB gene, 3 COX genes, 3 ATP genes, 1 ribosomal protein gene, and 15 open reading frames (ORF236, ORF458, ORF484, ORF477, ORF495, ORF408, ORF853, ORF838, ORF922, ORF371, ORF398, ORF227, ORF314, ORF410, and ORF195). A large number of ORFs showed unknown functions. Furthermore, the overall basic composition of M. fructicola was as follows: T: 33.7%, C: 14.2%, A: 35.0%, and G: 17.1%, and the content of GC is 31% with A + T-rich feature.
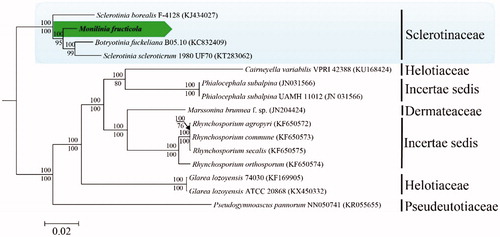
In this study, phylogenetic analysis was conducted with the complete mitogenomes of 14 relative species downloaded from NCBI and M. fructicola sequenced. The multiple mitogenomic sequences were aligned by HomBlocks (Bi et al. Citation2018), resulting in alignments with 5978 bp. The phylogenetic tree was constructed by Bayesian inference (BI) and maximum likelihood (ML) methods, and ML analysis which adopted GTR + G + I model was implemented in the soft RAxML version 7.0.3 (Stamatakis Citation2014), with 1000 bootstrap replicates. BI analysis applied MrBayes v.3.1.2 (Ronquist et al. Citation2012) for three million generations with the most appropriate model. Phylogenetic trees are displayed in . This figure shows that the results of BI analysis are consistent with that of ML analysis from 15 species of Leotiomycetes. The phylogenetic tree showed the structure of Leotiomycetes and classified 14 species into two incertae sedis and four families, including Sclerotiniaceae, Helotiaceae, Dermateaceae and Pseudeurotiaceae (). Meanwhile, M. fructicola was clustered in Sclerotiniaceae family and was tightly clustered in Botryotinia fuckeliana and Sclerotinia scleroticrum.
Figure 1. Phylogenetic analysis of mitochondrial genomes from Monilinia fructicola and its related species in Helotiales based on Bayesian inference and Maximum likelihood. Bayesian posterior probabilities and ML bootstrap values are shown above and below the internodes, respectively. GenBank accession numbers are shown in parentheses.
Declaration statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Choi IY, Kim J, Seo KW, Oh HT, Cho CH, Kim JH, Song YJ. 2016. Occurrence of brown rot on apricot caused by Monilinia fructicola in Korea. Res Plant Dis. 22:122–126.
- EFSA Panel on Plant Health (PLH). 2011. Pest risk assessment of Monilinia fructicola for the EU territory and identification and evaluation of risk management options. EFSA J. 9:2119.
- Grzegorczyk M, Żarowska B, Restuccia C, Cirvilleri G. 2017. Postharvest biocontrol ability of killer yeasts against Monilinia fructigena and Monilinia fructicola on stone fruit. Food Microbiol. 61:93–101.
- Hu MJ, Cox KD, Schnabel G, Luo CX. 2011. Monilinia species causing brown rot of peach in China. PLoS One. 6:e24990.
- Martini C, Mari M. 2014. Monilinia fructicola, Monilinia laxa (Monilinia Rot, Brown Rot). In: Bautista-Banos S, editor. Postharvest decay. Control strategies. Eastbourne (UK): Academic Press, Elsevier; p.233–258.
- Oliveira LL, Pacheco I, Mercier V, Faoro F, Bassi D, Bonard I, Quilot-Turion B. 2016. Brown rot strikes Prunus fruit: an ancient fight almost always lost. J Agric Food Chem. 64:4029–4047.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tran TT, Li H, Nguyen DQ, Sivasithamparam K, Jones MGK, Wylie SJ. 2017. Spatial distribution of Monilinia fructicola and M. laxa, in stone fruit production areas in Western Australia. Australasian Plant Pathol. 46:339–349.
