Abstract
Clematis is an important horticultural and medicinal genus in Ranunculaceae. Here, we report three complete chloroplast genome sequences of C. acerifolia, C. smilacifolia, and C. uncinata. The chloroplast genomes were found to be 159,552 bp, 159,624 bp, and 159,524 bp in length, and GC contents were 37.9, 37.9, and 38.0%, respectively. The sequence of each species contained 112 unique genes, including 29 tRNA, 4 rRNA, and 79 protein-coding genes. These chloroplast genome sequences provide knowledge to the genetic variations and evolution of Clematis and Ranunculaceae.
Clematis is one of the largest genera in the buttercup family (Ranunculaceae) with about 300 species distributed worldwide (Tamura Citation1995; Johnson Citation1997; Wang and Li Citation2005). Many Clematis species are traditional chinese medicines (TCM) and their garden hybrids are popular to gardeners. Here, we report three complete chloroplast genome sequences of Clematis. Among them, C. acerifolia is an endangered species narrowly endemic to Beijing (López-Pujol et al. Citation2005), C. uncinata is an important TCM species (Li et al. Citation2014), and C. smilacifolia is rare in southern China (Wang Citation2006).
Fresh young leaves of Clematis acerifolia, C. smilacifolia, and C. uncinata were collected from Shidu, Beijing (N 39°38′51″, E 115°35′58″), Longzhou, Guangxi (N 22°18′57″, E 106°55′24″, and Guilin, Guanxi (N 25°13′52″, E 110°12′31″, respectively. Voucher specimens were deposited in the Herbarium of Beijing Forestry University (BJFC) (under collection numbers of L. Xie 2012008, L. Xie 20151206, and L. Xie 20151171). Genomic DNAs were extracted using CTAB method (Doyle and Doyle Citation1987), and 2 × 150 bp pair end sequencing was performed on an Illumina HiSeq 4000 platform at Novogene (http://www.novogene.com, China). We used map function of Geneious R11 (Kearse et al. Citation2012) to extract chloroplast reads using published chloroplast genome from Clematis as references. Filtered chloroplast reads were used for de novo assembly with Geneious R11. Gaps were bridged using fine tuning function of Genious R11. The three chloroplast sequences were then annotated using Plann (Huang and Cronk Citation2015) and annotations were verified by Genious R11.
The plastome of C. acerifolia was 159,552 bp in length containing a large single-copy (LSC) region of 79,315 bp, a small single-copy (SSC) region of 18,145 bp, and two inverted repeats (IR) of 31,046 bp. The genome contained 112 functional genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. The sequence GC content was 37.9%. The length of plastome sequence of C. smilacifolia was 159,624 bp, which consisted of contained LSC region of 79,398 bp, SSC region of 18,125 bp, and IR regions of 31,050 bp. Gene numbers and GC content were detected in the same way as C. acerifolia. In C. uncinata, the plastome sequence was 159,524 bp in length. The LSC, SSC, and IR regions were 79,349 bp, 18,099 bp, and 31,038 bp, respectively. The gene numbers were the same with the other two tested species and the GC content was 38.0%. Gene inversions, transpositions, and IR expansion of the three samples were consistent with (Liu, Ding et al. Citation2018; Liu, He et al. Citation2018).
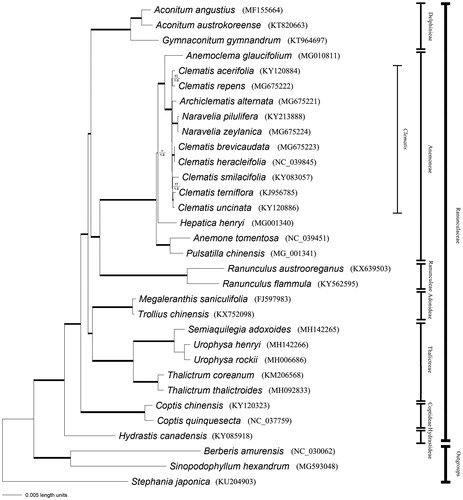
Complete chloroplast genome sequences of the other genera of Ranunculaceae available from GenBank were mined for phylogenomic analysis. Bayesian inference and parsimony analyses were conducted for phylogeny reconstruction (). The sequence alignment and all the settings of parsimony and Bayesian analyses were the same with Liu, Ding et al. Citation2018a. Phylogenetic framework of Clematis as well as Ranunculaceae were consistent with previous studies (Miikeda et al. Citation2006; Wang et al. Citation2009; Xie et al. Citation2011; Lehtonen et al. Citation2016; Jiang et al. Citation2017; Liu, Ding et al. Citation2018; Liu, He et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026.
- Jiang N, Zhou Z, Yang JB, Zhang SD, Guan KY, Tan YH, Yu WB. 2017. Phylogenetic reassessment of tribe Anemoneae (Ranunculaceae): nonmonophyly of Anemone s. l. revealed by plastid datasets. PLoS One. 12:e0174792.
- Johnson M. 1997. Släktet Klematis. Södertälje: M. Johnsons plantskola AB; p. 2–23.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lehtonen S, Christenhusz MJM, Falck D. 2016. Sensitive phylogenetics of Clematis and its position in Ranunculaceae. Bot J Linn Soc. 182:825–867.
- Li SG, Huang XJ, Li MM, Wang M, Feng RB, Zhang W, Li YL, Wang Y, Ye WC, 2014. Triterpenoid saponins from the roots of Clematis uncinata. Chemical and Pharmaceutical Bulletin. 62:35–44.
- Liu HJ, Ding CH, He J, Cheng J, Pei LY, Xie L. 2018. Complete chloroplast genomes of Archiclematis, Naravelia and Clematis (Ranunculaceae), and their phylogenetic implications. Phytotaxa. 343:214–226.
- Liu HJ, He J, Ding CH, Lyu RD, Pei LY, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Pl Sci. 9:1097.
- López-Pujol J, Zhang FM, Ge S. 2005. Population genetics and conservation of the critically endangered Clematis acerifolia (Ranunculaceae). Can J Bot. 83:1248–1256.
- Miikeda O, Kita K, Handa T, Yukawa T. 2006. Phylogenetic relationships of Clematis (Ranunculaceae) based on chloroplast and nuclear DNA sequences. Bot J Linn Soc. 152:153–168.
- Tamura M. 1995. Clematis L. In: Heipko P, editor. Engler’s Die Natürlichen Pflanzenfamilien. 2nd ed. Berlin: Duncker & Humblot; p. 368–387.
- Wang W, Lu AM, Ren Y, Endress ME, Chen ZD. 2009. Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspect Pl Eco Evol Syst. 11:81–110.
- Wang WT. 2006. A revision of Clematis sect. Naraveliopsis (Ranunculaceae). Acta Phytotax Sin. 44:670–699.
- Wang WT, Li LQ. 2005. A new system of classification of the genus Clematis (Ranunculaceae). Acta Phytotax Sin. 43:431–488.
- Xie L, Wen J, Li LQ. 2011. Phylogenetic analyses of Clematis (Ranunculaceae) based on sequences of nuclear ribosomal ITS and three plastid regions. Syst Bot. 36:907–921.