Abstract
To study the intraspecific variation of the sea cucumber, Apostichopus japonicus, we sequenced three complete mitochondrial genomes, especially from different body color variants (green, black, and red) of Apostichopus japonicus in Geomum-do, Republic of Korea. The full lengths of green, black, and red sea cucumber mitochondrial genomes are 16,106 bp, 16,096 bp, and 16,107 bp, respectively. They all contain 37 genes (13 protein-coding genes, 2 rRNAs, and 22 tRNAs), and have an average 38.0% of GC ratio. Two hundred thirty eight single nucleotide polymorphisms and 26 insertion and deletions were identified among three sea cucumber mitochondrial genomes. The phylogenetic analysis of A. japonicus with additional 14 available A. japonicus and two Parastichopus mitochondrial genomes were used as outgroups shows somewhat distinct three clades with body color patterns. Furthermore, there is no clear phylogenetic separation among Korean, Chinese, and Japanese individuals. These data will provide new insights and to facilitate the rapid growth of the sea cucumber aquaculture industry and its breeding programs.
The sea cucumber Apostichopus japonicus (Selenka 1867) is one of the economically important farmed species as a source of nutritional food and traditional medical resource widely and increasingly consumed in northeast Asia. Intriguingly, there are three major body color variants with different biological and morphological characteristics (Yang et al. Citation2015). To establish a sustainable breeding strategy for sea cucumber aquaculture, it is necessary to elucidate the taxonomic status of these body color variants.
We obtained genomic DNA from red, green, and black wild specimens of A. japonicus in the same geographic region, near the coast of Geomun-do in South Korea (34°1′35′′N, 127°18′45′′E) using a DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). The remaining cells and samples were deposited at the Wildlife Specimen Bank in Chonnam National University, Korea. Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Seoul, Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008). All bases were confirmed by alignment results generated by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation.
The three mitochondrial genomes from green, black, and red body color of A. japonicus, named as KG, KB, and KR, (Genbank accessions are MK208925, MK216560, and MK216561, respectively) are 16,106 bp, 16,096 bp, and 16,107 bp long, which is similar to the 14 available other A. japonicus mitochondrial genomes in length (16,096 bp–16,109 bp). GC ratio of the three mitochondrial genomes are 38.0, 38.1, and 38.1%, respectively and all three genomes contain 13 protein-coding genes, two rRNAs, and 22 tRNAs with the same order. Among KG, KR, and KB complete mitochondrial genomes, a total of 238 SNPs and 26 INDELs are identified: 143 SNPs and 7 INDELs (56.8%) of KG-specific, 58 SNPs and 8 INDELs (25.0%) of KB-specific, and 40 SNPs and 4 INDELs (16.7%) of KR-specific.
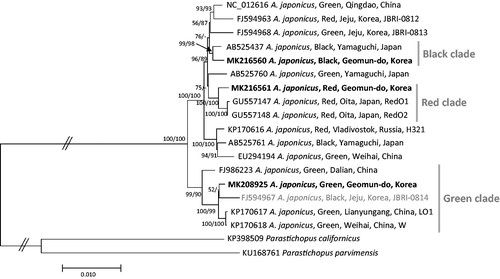
Seventeen complete mitochondrial genomes of sea cucumbers (Sun et al. Citation2010; Zhang, Cao et al. Citation2016) and those of outgroup species, Parastichopus californicus (KP398509; Zhang, Bao et al. Citation2016) and Parastichopus parvimensis (KU168761) were used to construct phylogenetic trees using maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (boostrap repeat is 10,000) methods. Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) and trees were generated by MEGA X (Kumar et al. Citation2018). Trees show that three clades representing three body colors of sea cucumbers cover 9 of 17 sea cucumbers; while eight sea cucumbers are not clustered by neither body color or its geographical origin (). Along with these results, there is no clear relationship between genetic background and both geographical distribution and body color of sea cucumber, which is the same to previous several studies (Sun et al. Citation2010; Jo et al. Citation2016; Zhang, Cao et al. Citation2016). These data will provide new insights and to facilitate the rapid growth of the sea cucumber aquaculture industry and its breeding programmes.
Figure 1. Maximum likelihood and neighbor joining phylogenetic trees of 19 mitochondrial genomes: six Korean sea cucumbers (FJ594963, FJ594967, FJ594968, MK208925, MK216560, and MK216561), five Chinese sea cucumbers (NC_012616, EU294194, FJ986223, KP170617, and KP170618), five Japanese sea cucumbers (AB525437, AB525760, GU557147, GU557148, and AB525761), one Russian sea cucumber (KP170616), Parastichopus californicus (KP398509), and Parastichopus parvimensis (KU168761). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining trees, respectively. Grey color on OTU name indiciates exceptional case of skin color specific clade.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jo J, Park C, Kim M, Park C. 2016. Phylogenetic analysis of the three color variations of the sea cucumber Apostichopus japonicus. J Aquac Res Development. 7:2.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Sun X-J, Li Q, Kong L-F. 2010. Comparative mitochondrial genomics within sea cucumber (Apostichopus japonicus): provide new insights into relationships among color variants. Aquaculture. 309:280–285.
- Yang H, Hamel J-F, Mercier A. 2015. The sea cucumber Apostichopus japonicus: history, biology and aquaculture. Vol. 39. UK: Academic Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang W, Cao Z, Li Y, Zhao H, Huang J, Liang Z, Huang L. 2016. Taxonomic status of the three color variants in sea cucumber (Apostichopus japonicus): evidence from mitochondrial phylogenomic analyses. Mitochondrial DNA A. 27:2330–2333.
- Zhang Z, Bao X, Dong Y, Gao X, Gao L, Li S, Liu W, Hou H, Shi J, Pu H. 2016. Complete mitochondrial genome of Parastichopus californicus (Aspidochirotida: Stichopodidae). Mitochondrial DNA A. 27:3569–3570.
