Abstract
The complete chloroplast genome of Elatostema dissectum Wedd. was reported in this study. The cp genome was 150,302 bp in length including two inverted repeats (IRs) of 24,565 bp, which were separated by LSC and SSC of 83,998 bp and 17,174 bp, respectively. The GC content was 36.2%. The genome encoded 112 functional genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. This plastid genome is the first report for the tribe Elatostemateae in Urticaceae, which will be useful data for developing markers for further studies on resolving the relationship within the genus and tribe.
Elatostema J. R. Forst. and G. Forst (Urticaceae) comprises more than 500 species of herbs and subshrubs that grow in deep shade forests, gorges, caves, and stream sides (Wang Citation2014; Fu, Monro et al. Citation2017). Elatostema is distributed in tropical and subtropical Africa, Australia, Asia, and Oceania. This genus has been revealed to represent an ideal model on the aspects of reproductive patterns in karst groups and for long-distance dispersal researches (Fu, Su et al. Citation2017; Wu, Liu et al. Citation2018). Despite Wu et al. (Citation2013) reconstructed the phylogenetic relationship of Urticaceae, which confirmed the monophyly of the tribe Elatostemateae, the relationships within Elatostema still remain poorly resolved.
Chloroplast genome provides additional information to resolve the phylogenetic relationships at generic and infra-generic level. Within Urticaceae, only three chloroplast genomes of two Boehmeria and one Cecropia species have been published (Wu et al. Citation2017; Wu, Du et al. Citation2018) to date; these genera belong to the tribes other than Elatostemateae.
In this study, leaf materials were collected from plants of Elatostema dissectum in the field of Jinxiu County, Guangxi, China (N 110°6′46″, E 23°58′22″) and immediately dried by silica gel for DNA extraction. Voucher specimen (XZB20180131-01) of this collection was deposited at IBK. Total genome DNA was extracted using a modified protocol (Chen et al. Citation2014) and sent to Novogene Company (http://www.novogene.com, China) for next generation sequencing using Illumina Hiseq 4000. We used map to reference function in Geneious 11.1.5 (Kearse et al. Citation2012) to exclude nuclear and mitochondrial reads using published plastid genome of Cecropia pachystachya (MF953831) as reference. Putative chloroplast reads were used for de novo assembling construction using Geneious 11.1.5. Generated contigs were annotated and concatenated into larger ones based on the reference of C. pachystachya using Geneious 11.1.5. The original data were again mapped to the larger contigs to extend their boundaries until all contigs were able to concatenate to one contig. The IR region was determined using the repeat finder function in Geneious 11.1.5 and was inverted and copied to construct the complete chloroplast sequence. The cp genome was manually adjusted to remove ambiguous sites. The annotation process was performed following Liu et al. (Citation2018) using Morus mongolica (KM491711) as the reference. The complete chloroplast genome of Elatostema dissectum was 150,302 bp in length (MK227819), the GC content was 36.2%. LSC and SSC contained 83,998 bp and 17,174 bp, respectively, while IR was 24,565 bp in length. The plastid genome encoded 112 functional genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes.
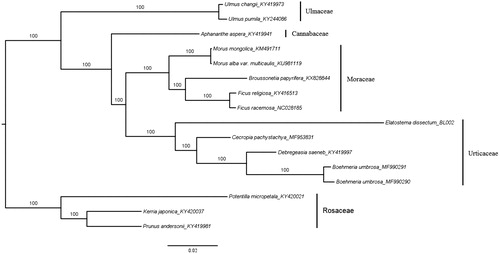
The maximum likelihood phylogenetic tree was reconstructed based on the complete genomes of Elatostema dissectum, four species from Urticaceae and additional 11 species from Cannabaceae, Moraceae, Rosaceae, and Ulmaceae (). The result was congruent with previous studies (Wu et al. Citation2013; Wu, Liu et al. Citation2018) and showed that E. dissectum representing tribe Elatostemateae is sister to the group of Boehmeria, Debregeasia, and Cecropia with strong support. The newly reported chloroplast genome of E. dissectum allows developing markers for further studies on resolving the relationship within the tribe Elatostemateae and particularly, the genus Elatostema.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen LY, Song MS, Zha HG, Li ZM. 2014. A modified protocol for plant genome DNA extraction. Plant Diversity and Resour. 36:375–380.
- Fu LF, Monro AK, Huang SL, Wen F, Wei YG. 2017. Elatostema tiechangense (Urticaceae), a new cave-dwelling species from Yunnan, China. Phytotaxa. 292:85–090.
- Fu LF, Su LY, Mallik A, Wen F, Wei YG. 2017. Cytology and sexuality of 11 species of Elatostema (Urticaceae) in limestone karsts suggests that apomixis is a recurring phenomenon. Nor J Bot. 35:251–256.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu H, He J, Ding C, Lyu R, Pei L, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Plant Sci. 9:1097.
- Wang WT. 2014. Elatostema (Urticaceae) in China. Qingdao: Qingdao Press.
- Wu ZY, Monro AK, Milne RI, Wang H, Yi TS, Liu J, Li DZ. 2013. Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Mol Phylogenet Evol. 69:814–827.
- Wu ZY, Du XY, Milne RI, Liu J, Li DZ. 2017. Charaterization of the complete chloroplast genome sequence of Cecropia pachystachya. Mitochondrial DNA B Resour. 2:735–737.
- Wu ZY, Liu J, Provan J, Wang H, Chen CJ, Cadotte MW, Luo YH, Amorim BS, Li DZ, Milne RI. 2018. Testing Darwin’s transoceanic dispersal hypothesis for the inland nettle family (Urticaceae). Ecol Lett. 21:1515–1529.
- Wu ZY, Du XY, Milne RI, Liu J, Li DZ. 2018. Complete chloroplast genome sequences of two Boehmeria species Urticaceae. Mitochondrial DNA B Resour. 3:939–940.