Abstract
Loxosceles rufescens, the Mediterranean recluse spider, is a cosmopolitan species with toxic venom which can cause skin lesions (arachnogenic necrosis) in humans. Here, the complete mitochondrial genome (mitogenome) was determined for L. rufescens (Sicariidae). It had a circular mapping organization with 15,210 bp in length containing 13 protein-coding genes, two ribosomal RNA genes, 21 putative transfer RNA genes, and one control region. A Bayesian phylogenomic analysis of the representative species in Araneae showed that L. rufescens had a closer relationship with two species of Pholcidae and one species of Hypochilidae, which were the haplogyne spiders. The complete mitogenome will aid identification of L. rufescens, facilitate further genetic studies of the significant pest, and contribute to spider phylogenetic inferences.
Loxosceles rufescens belongs to the family Sicariidae of Haplogynae in Araneae (World Spider Catalog Citation2018). This species (as many haplogyne spiders) is poor in distinctive pattern, especially since it has rather simple sexual organs (Brignoli Citation1969, Citation1976). Recent studies (Luo and Li Citation2015; Nentwig et al. Citation2017) have found that L. rufescens is a cryptic species complex. Therefore, the identification of L. rufescens and separation from similar species is not easy based on morphology (see Greene et al. Citation2009; Vetter et al. Citation2015). The complete mitochondrial genome (mitogenome) is useful for molecular identification of this species, studies of its population genetics, and inference of phylogenetic relationships among related taxa. In this article, we sequenced the complete mitogenome of L. rufescens, and conducted phylogenetic analysis of the representative species in Araneae using 13 mitochondrial protein-coding genes.
Loxosceles rufescens was collected in a cave in Taihe, Jiangxi, China (26°31′N, 115°07′W). The specimen and DNA were deposited in the Institute of Zoology, Chinese Academy of Sciences (IZCAS), Beijing, China. Total genomic DNA was isolated using a TIANamp Micro DNA Kit (Tiangen Biotech CO., LTD) following the manufacturer’s Instructions. Primers designed to match generally conserved regions of target mitochondrial DNA were used to amplify short fragments from 16s, 12s, cox1, cox3, ND5, CYTB. Specific primers were designed based on these conserved regions sequences and used to amplify the remained mitochondrial DNA sequences in several PCR reactions. Every PCR solution (50 μL) contained 0.2–1.0 μmol of each primer, 2.5 U TaKaRa LATaq (Takara), 5 μL 10 × PCR buffer, 10 mmol deoxyribonucleotide triphos-phate (dNTP) mixture (2.5 mmol of each dNTP) and 60 ng of total DNA. The PCR reaction was carried out with LATaq polymerase for 35 cycles at 94 °C for 30 s, and annealed at 50 °C for 30 s, followed by extension at 72 °C for 1 min per 1 kb. The final MgCl2 concentration in the PCR reaction was 2.0 mmol/L. PCR products were cloned into pMD18-T vector (Takara, JAP) and then sequenced, or sequenced directly by the dideoxynucleotide procedure, using an ABI 3730 automatic sequencer. Sequences were assembled by software of DNAstar and adjusted manually to generate the complete sequence of mitochondrial DNA.
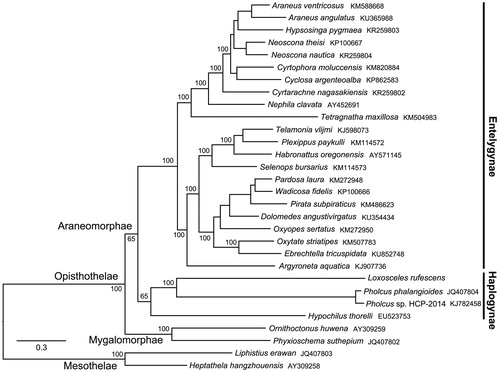
The complete mitogenome of L. rufescens is a circular molecule of 15,210 bp (GenBank accession number MK257773). It consists of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 21 putative transfer RNA (tRNA) genes, and one control region. In addition, 8 intergenic spacers (103 bp in total) and 21 overlapping sequences (227 bp in total) are interspersed throughout the genome. The overall A + T content of mitogenome is 63.8% (A = 23.27%, T = 36.53%, G = 25.58%, C = 10.62%). The putative control region (1737 bp), located between tRNA-Met and tRNA-Gln, had an A + T bias of 70.3%. Thirteen PCGs were extracted from mitogenomes of 30 spider species to reconstruct the Bayesian phylogeny (). A phylogenomic analysis showed the 26 species of Araneomorphae were divided into two groups: Haplogynae and Entelegynae. L. rufescens together with reported Pholcus phalangioides, Pholcus sp. HCP-2014 and Hypochilus thorelli were all in Haplogynae clade, and the remaining 22 species formed a monophyletic Entelegynae group.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Brignoli PM. 1969. Note sugli Scytodidae d’Italia e Malta (Araneae). Fragm Entomol. 6:121–166.
- Brignoli PM. 1976. Beiträge zur Kenntnis der Scytodidae (Araneae). Revue Suisse Zool. 83:125–191.
- Greene A, Breisch NL, Boardman T, Pagac BB, Jr Kunickis E, Howes RK, Brown PV. 2009. The Mediterranean recluse spider, Loxosceles rufescens (Dufour): an abundant but cryptic inhabitant of deep infrastructure in the Washington, D.C. area (Arachnida: araneae: Sicariidae). Am Entomol. 55:159–167.
- Luo Y, Li S. 2015. Global invasion history of the Mediterranean recluse spider: a concordance with human expansion. Ecography. 38:1080–1089.
- Nentwig W, Pantini P, Vetter RS. 2017. Distribution and medical aspects of Loxosceles rufescens, one of the most invasive spiders of the world (Araneae: Sicariidae). Toxicon. 132:19–28.
- Vetter RS, Swanson DL, Weinstein SA, White J. 2015. Do spiders vector bacteria during bites? The evidence indicates otherwise. Toxicon. 93:171–174.
- World Spider Catalog. 2018. World Spider Catalog. Version 19.5. Natural History Museum Bern, online at http://wsc.nmbe.ch, accessed on 21 October 2018.