Abstract
The complete mitochondrial genome (mitogenome) of Euchorthippus unicolor was sequenced for the first time with a length of 15,629 bp. The base nucleotide composition in the mitogenome of E. unicolor was: A(43.3%), T(32.8%), C(13.7%), G(10.2%), and AT(76.1%), respectively. All protein-coding genes start with typical ATN codon except cox1, which initiates with CCG codon instead and ends with either complete TAA/TAG codons or incomplete TA(a) codons. The phylogenetic analysis indicated that Euchorthippus unicolor was clustered with genus Gomphocerus species.
Euchorthippus unicolor is a grasshopper belonging to the Euchorthippus Tarb, Arcypterinae, Arcypteridae, Acridoidea, Orthoptera (Zheng and Xia Citation1998), which has a closer relationship with Chorithippus fieber. The Euchorthippus Tarb contained 26 species, distributed in Europe and Asia. In the present study, we sequenced and analysed the mitogenome of E. unicolor, with the number of GenBank, MK113716. The sample of E. unicolor was collected from the small South Moutain (Xi’an, China) in 2013 and was deposited in the Molecular and Evolutionary Lab in Shaanxi Normal University in China. The leg muscle of the grasshopper was used for the total genomic DNA isolation and the orthopteran universal mitochondrial primers were performed following the study of Liu et al. (Citation2006) and Simon et al. (Citation2006). The mitogenome sequence of E. unicolor was assembled and annotated with the software of the Staden Package 1.7 (Staden et al. Citation2000).
The complete mitogenome of E. unicolor was determined to be a length of 15,629 bp, comprising 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and one A + T-rich region. The tRNA genes were predicted by the online software tRNAScan-SE (Lowe and Eddy Citation1997) and the length ranged from 63 bp (tRNA-Thr) to 71 bp (tRNA-Val and tRNA-Lys). The length of 12S rRNA were 828 bp and 16S rRNA was 1319 bp, located between tRNA-Leu (CUN) and A + T-rich region. All protein coding genes started with the typical start codon ATN, except cox1 which started with CCG. All protein coding genes end with TAA or TAG, while nad5 ends with the incomplete stop codon TA. The A + T-rich region has a length of 738 bp. The base nucleotide composition in the mitogenome of E. unicolor was: A(43.3%), T(32.8%), C(13.7%), G(10.2%), and AT(76.1%), respectively, showing an obvious AT bias as appearing in the other orthopteran species (Qiu et al. Citation2017).
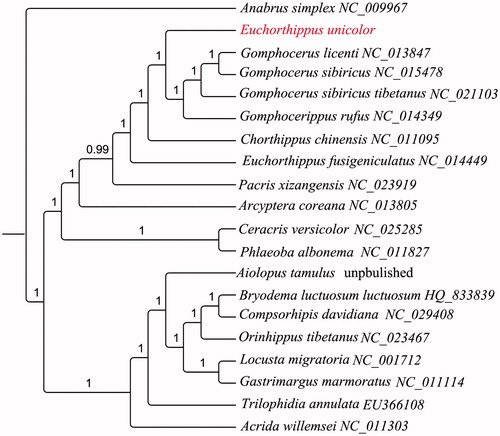
To conform the phylogenetic relationship of E. unicolor, a phylogenetic tree was built in the superfamily Acridoidea based on 13 PCGs with 19 ingroup and 1 outgroup using MrBayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) (). The result showed that E. unicolor was clustered together with genus Gomphocerus.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Liu N, Hu J, Huang Y. 2006. Amplification of grasshoppers complete mitochondrial genomes using long PCR. Chin J Zool. 41:61–65. (in Chinese with an English abstract).
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Qiu ZY, Yuan H, Wang XL, Cui YY, Lian T, Mao SL. 2017. Characterization of the mitochondrial genome of Phyllomimus sp. (Orthoptera: Pseudophyllidae). Mitochondrial DNA Part B. 2:900–901.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Staden R, Beal KF, Bonfield JK. 2000. The Staden package, 1998. Methods Mol Biol. 132:115–130.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach A. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(37):545–579.
- Zheng ZM, Xia KL. 1998. Orthoptera: Acridoidea: Oedipodidae and Arcypteridae. Fauna Sinica Insecta. Vol.10. Beijing: SciencePress; pp.216–539.