Abstract
The complete mitochondrial genome of Cryptophyceae sp. CCMP2293 was 42,262 bp and contained 41 protein-coding genes (PCG), 2 ribosomal RNA (rRNA), and 26 transfer RNA (tRNA) genes that are typical of cryptophyte algae mtDNA. A phylogenetic analysis based on the mitochondrial genomes of cryptophyte algae indicated that Cryptophyceae sp. CCMP2293 and Teleaulax amphioxeia strain HACCP-CR01 were the most closely related species, which strongly supports their close phylogenetic affinity.
Cryptophyte algae are an ecologically important group that consists of heterotrophic, phototrophic, and osmotrophic species inhabiting marine and freshwater environments (Graham and Wilcox Citation2000; Shalchian-Tabrizi et al. Citation2008). Evolutionary biologists are greatly interested in these species because their plastids are of red algal secondary endosymbiotic origin (Mcfadden Citation1993). These algae have two flagella with hairs and a dorsiventral shape with the cells flattened on one plane, and they contain chlorophyll a, chlorophyll c, beta carotene, and phycobilins as accessory pigments. Cryptophytes have strong adaptability to light and temperature, and they become the dominant species in both winter and summer. Most cryptophytes contain four distinct genomes: nuclear, nucleomorph, mitochondrial, and plastid genomes (Douglas et al. Citation1991). Therefore, cryptophytes are considered an interesting model for studying genome evolution and protein targeting.
In our study, the species of Cryptophyceae sp. CCMP2293 was provided by the Culture Collection of Marine at the Ocean University of China in Qingdao (OUC- 2013060210). The complete mitochondrial genome of Cryptophyceae sp. CCMP2293 is sequenced using PCR with 26 pairs of primers. The complete genome of Cryptophyceae sp. CCMP2293 was 42,262 bp in length and contained 41 PCGs (atp1, atp4, atp6, atp8-9, rps1-4, rps7-8, rps11-14, rps19, cox1-3, nad1-11, nad4L, rpl5-6, rpl14, rpl16, rpl31, ORF167, sdh3-4, tatC, and cob), 26 tRNA genes, 2 ribosomal RNA (rRNA) genes. 38 PCGs used ATG as the typical initiation codon, and 3 PCGs (atp 1, rps2, and rps12) used TTG as typical initiation codons. In total, 32, 5, and 4 PCGs had the complete termination codons TAA, TAG, and TGA, respectively. No incomplete stop codons were found. The overall GC content was 30.36%, which is well within the normal range of cryptophyte algae mitochondrial DNA. All 26 typical tRNAs, which ranged from 70 to 86, possessed a complete cloverleaf secondary structure.
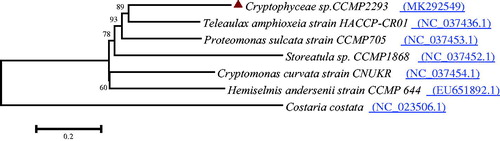
To determine the phylogenetic position of Cryptophyceae sp. CCMP2293 in Cryptophyceae, 6 complete mitochondrial genome sequences were obtained from the Genbank database. Phylogenetic analysis using the maximum likelihood method implemented in MEGA6 (Tamura et al. Citation2013) based on 21 PCGs showed that Cryptophyceae sp. CCMP2293 is the closest sister species to Teleaulax amphioxeia strain HACCP-CR01 ().
The complete mitochondrial genome sequence obtained in our study would be useful for studying the phylogenetic history of Cryptophyceae sp. CCMP2293 and related species.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Douglas SE, Murphy CA, Spencer DF, Gray MW. 1991. Cryptomonad algae are evolutionary chimaeras of two phylogenetically distinct unicellular eukaryotes. Nature. 350:148–151.
- Graham LK, Wilcox LW. 2000. The origin of alternation of generations in land plants: a focus on matrotrophy and hexose transport. Philo Transact: Biol Sci. 355:757–767.
- Mcfadden GI. 1993. Second-hand chloroplasts: evolution of cryptomonad algae. Adv in Bot Res. 19:189–230.
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. 2008. Multigene phylogeny of Choanozoa and the origin of animals. PLoS One. 3:e2098
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729.