Abstract
We have determined the mitochondrial genome of Chilo suppressalis (Walker, 1863) collected in Korea. The circular mitogenome of C. suppressalis was 15,341 bp including 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 622 bp. The base composition was AT-biased (93.6%). Through alignment of three C. suppresalis mitochondrial genomes, 79 single nucleotide polymorphisms (8.8%) and 291 insertions and deletions (33.3%) are identified as Korean C. suppressalis specific variations. Phylogenetic trees present that three mitochondrial genomes of C. suppressalis have enough variations within a species, which can be a candidate to understand its origin and geographical distribution using molecular marker sequences.
Chilo suppressalis (Walker, 1863), the striped stem borer, is one of the rice pests in Eastern and Southeastern Asia including China, Korea, India, and Indonesia (Park and Hyun Citation1990; Meng et al. Citation2008). Because of its ecological importance in agriculture (Pathak and Khan Citation1994), biological agents or sex pheromones have been developed (Beevor et al. Citation1990; Harry Citation1991; Ye et al. Citation2001). Till now, two complete mitochondrial genomes of C. suppressalis collected in China (Yin et al. Citation2011; Chai et al. Citation2012) were sequenced, which is a good resource to understand genetic diversity of C. suppressalis.
To understand genetic differences of C. suppressalis between Korea and China, we collected C. suppressalis in Taean-gun, South Chungcheong Province, Korea (GPS coordination is N36.875787, E126.227316). The specimens and DNA were deposited in the Insect Collection of Gyeongsang National University, South Gyeongsang Province, Korea (GNU#170303-001). Total DNA was extracted from C. suppressalis and genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea. de novo assembly was conducted by Velvet 1.2.10 (Zerbino and Birney Citation2008) and gaps were filled by SOAPGapCloser 1.12 (Zhao et al. Citation2011). All bases were confirmed by alignments against assembled sequences generated by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li H et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for mitochondrial genome annotation using C. suppressalis complete mitochondrial genome (NC_015612).
The mitochondrial genome of C. suppressalis (Genbank accession is MK207057) is 15,341 bp in length. The nucleotide composition is AT-biased (79.6%). The mitogenome contains 13 protein-coding genes, two rRNAs, and 22 tRNAs. The tRNAs size ranges from 64 to 76 bp, are similar to other insects (circa 54–90 bp). Gene order of C. suppressalis mitochondrial genome is almost same to those of 22 complete mitochondrial genomes in Crambidae except that (i) tRNA-Val found between two rRNAs in four genomes, and (ii) cytochrome B enzyme gene was missing in Spoladea recurvalis (NC_027443; He et al. Citation2015). Their genome lengths range from 15,110 bp (Maruca testulalis; NC_024283; Zou et al. Citation2016) to 15,490 bp (Diatraea saccharalis; NC_013274; Li W et al. Citation2011): that of C.suppressalis is near to the longest. The control region presumably corresponds to the single largest non-coding AT-rich region (348 bp, A + T 95.4%).
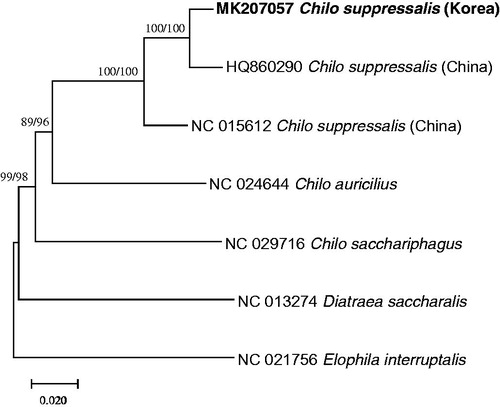
Seven complete mitochondrial genomes from Crambidae including two mitochondrial genomes of C. suppressalis were aligned by ClustalW (Thompson et al. Citation2003) for constructing phylogenetic trees using maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) methods using MEGA X (Kumar et al. Citation2018). From the alignment of three C. suppressalis complete mitochondrial genomes, 894 single nucleotide polymorphisms (SNPs) and 873 insertions and deletions (INDELs) are identified and among them, 79 SNPs (8.8%) and 291 INDELs (33.3%) are specific to Korean C. suppressalis. The phylogenetic trees supported by high bootstrap values present that three C. suppressalis mitochondrial genomes have enough variations within species (). Together with these results, C. suppressalis can be a candidate to understand its origin as well as a geographical distribution based molecular marker sequences.
Figure 1. maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees based on seven complete mitochondrial genomes in Crambidae: Korean C. suppressalis (MK207057, this study), Chinese C. suppressalis (NC_015612 and HQ860290), Chilo sacchariphagus (NC_029716), Chio auricilius (NC_024644), Diatraea saccharalis (NC_013274), and Elophila interruptalis (NC_021756). The numbers above the branches indicate bootstrap support values of maximum likelihood and neighbor joining trees.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Beevor P, David H, Jones O. 1990. Female sex pheromones of Chilo spp.(Lepidoptera: Pyralidae) and their development in pest control applications. Int J Trop Insect Sci. 11:785–794.
- Chai H-N, Du Y-Z, Zhai B-P. 2012. Characterization of the complete mitochondrial genomes of Cnaphalocrocis medinalis and Chilo suppressalis (Lepidoptera: Pyralidae). Int J Biol Sci. 8:561.
- Harry KK. 1991. Evaluation of entomopathogenic nematodes, Steinernema carpocapsae (Steinernematidae) and Heterorhabditis bacteriophora (Hetrorhabditidae) against Rice Stem Borer Chilo suppressalis (Walker)(Lepidoptera: Pyralidae). Kor J App Entomol. 30:50–53.
- He S-L, Zou Y, Zhang L-F, Ma W-Q, Zhang X-Y, Yue B-S. 2015. The complete mitochondrial genome of the beet webworm, Spoladea recurvalis (Lepidoptera: Crambidae) and its phylogenetic implications. PLoS One. 10:e0129355
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Li W, Zhang X, Fan Z, Yue B, Huang F, King E, Ran J. 2011. Structural characteristics and phylogenetic analysis of the mitochondrial genome of the sugarcane borer, Diatraea saccharalis (Lepidoptera: Crambidae). DNA and Cell Biology. 30:3–8.
- Meng X-F, Shi M, Chen X-X. 2008. Population genetic structure of Chilo suppressalis (Walker) (Lepidoptera: Crambidae): strong subdivision in China inferred from microsatellite markers and mtDNA gene sequences. Mol Ecol. 17:2880–2897.
- Park C, Hyun J. 1990. Studies on the regional characteristics in the occurrence of the rice stem borer, Chilo suppressalis Walker in Korea. Kor J App Entomol. 29:257–268.
- Pathak MD, Khan ZR. 1994. Insect pests of rice. International Rice Research Institute.
- Thompson JD, Gibson TJ, Higgins DG. 2003. Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics. 2.3. 1-2.3. 22.
- Ye G-Y, Shu Q-Y, Yao H-W, Cui H-R, Cheng X-Y, Hu C, Xia Y-W, Gao M-W, Altosaar I. 2001. Field evaluation of resistance of transgenic rice containing a synthetic cry1Ab gene from Bacillus thuringiensis Berliner to two stem borers. J Eco Entomol. 94:271–276.
- Yin J, Wang A-M, Hong G-Y, Cao Y-Z, Wei Z-J. 2011. Complete mitochondrial genome of Chilo suppressalis (Walker) (Lepidoptera: Crambidae)). Mitochondrial DNA. 22:41–43.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zou Y, Ma W, Zhang L, He S, Zhang X, Tao Z. 2016. The complete mitochondrial genome of the bean pod borer, Maruca testulalis (Lepidoptera: Crambidae: Spilomelinae). Mitochondrial DNA Part A. 27:740–741.
