Abstract
The complete mitochondrial genome of Promalactis odaiensis is described in this study. The mitogenome is 15,733 base pairs with 13 protein coding genes (PCGs), 2 rRNAs, 22 tRNAs, and a 679 bp AT-rich region. The overall base composition was 79.43% AT and 20.57% GC. Among 13 PCGs, eleven genes (Nad2, Atp8, Atp6, Cox3, Nad3, Nad5, Nad4, Nad4L, Nad6, CYTB, and Nad1) harbour the typical stop codon TAA or TAG, whereas remaining two genes (Cox1, Cox2) terminated with T.
Oecophoridae (Lepidoptera: Gelechioidea) is one of the most species-rich microlepidopteran family with worldwide distribution (Kim et al. Citation2016). It comprises more than 3300 described species (Nieukerken et al. Citation2011), and the highest diversity is in Australia with over 3000 endemic species (Common Citation2000). Unusually, Promalactis Meyrick is the Oriental and Palaearctic genus, comprising more than 275 species (Kim et al. Citation2018). Among them, the species, Promalactis odaiensis was reported by Park in 1980, and widely distributed from Eastern Asia including Korea, Japan and China (Kim and Lee Citation2016).
The individual of P. odaiensis was collected from Hangye-ri, Inje-gun, province Gangwon of South Korea (38°7′21.91″N, 128°18′8.09″E) on 9th August of 2018 by the first author. The genomic DNA was extracted by Exgene Tissue SV kit (GeneAll, Korea), and stored in our laboratory (Lab. of insect biosystematics, Seoul National University, South Korea). Illumina DNA libraries were constructed using the NEXTflex Rapid DNA-seq kit (Bioo Scientific Cor. Austin, TX, USA). High-throughput sequencing used the Illumina HiSeq 2500 platform (Illumina Inc., Sang Diego, CA, USA) at the Genome Analysis center of National Instrumentation Center for Environmental Management, Korea. The run mode was Rapid Pair End 250 cycles. A total 18,536,796 reads were analyzed to generate 4,652,735,796 base pairs of sequence and assembled in CLC genome assembler (ver. 4.010.83348, CLC Inc., Aarhus, Denmark). Annotation of protein-coding genes (PCGs), ribosomal RNAs (rRNAs), and transfer RNA (tRNA) genes were conducted using Ge-seq (Tillich et al. Citation2017). The complete mitogenome of P. odaiensis (15,733bp), which consisted of 37 individual genes, 13 PCGs, 2 rRNAs, 22 tRNA genes and A + T region. It was larger than those of other sequenced gelechioids with a range from 15,131 bp of Perimede sp. to 15,456 bp of Stathmopoda auriferella (Jeong et al. Citation2016). The 13 PCGs are similar to other insects, and 11 genes begin with typical ATN codon: three (Nad2, Nad3, and Nad6) start with ATA, one (Atp8) with ATC, six (Atp6, Cox3, Nad4, Nad4L, CYTB, and Nad1) with ATG and one (Nad5) with ATT, whereas Cox1 and Cox2 began with CGA and GTG, respectively. Among 13 PCGs, 11 genes (Nad2, Apt8, Atp6, Cox3, Nad3, Nad5, Nad4, Nad4L, Nad6, CYTB, and Nad1) harbour the typical stop codon, TAA or TAG, whereas remaining Cox1 and Cox2 terminate with T. The 679bp A + T rich region displayed as several lepidopteran-specific features, and located between 12S rRNA and tRNAMet. The overall base composition was 79.43% AT and 20.57% GC.
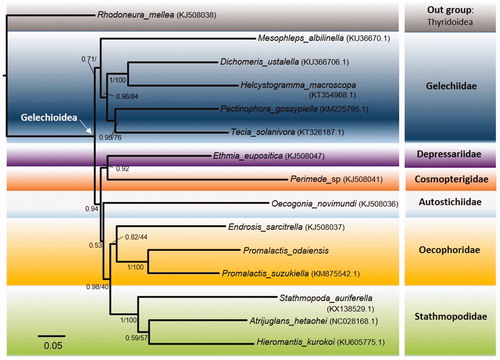
Using one species of Thyridoidea as an outgroup, the phylogeny of fourteen Gelechioidea species including P. odaiensis, was reconstructed based on nucleotide sequence data of 13 PCGs with Bayesian inference (BI) and maximum-likelihood (ML) analysis through MrBayes (ver. 3.1.2) and RAxML (CIPRES ver. 3.3) (). The phylogenetic position of P. odaiensis was closely clustered with P. suzukiella and Endrosis sarcitrella, and consisted of Oecophoridae which shows a monophyletic. Other families comprising more than two species, such as Gelechiidae and Stathmopodidae, were also shown as monophyletic.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of Promalactis odaiensis has been assigned GenBank accession number, MK210633.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Common IFB. 2000. Oecophorine genera of Australia III: the Barea group and unplaced genera (Lepidoptera: Oecophoridae). Monographs on Australian Lepidoptera, vol. 8. Melborune, Australia: CSIRO Publishing, pp.1–453.
- Jeong SY, Park JS, Kim SS, Kim I. 2016. Complete mitochondrial genome of the gelechioid Stathmopoda auriferella (Lepidoptera: Stathmopodidae). Mitochondrial DNA Part B: Res. 1:522–524.
- Kim S, Bae YS, Lee S. 2018. Genus Promalactis Meyrick (Lepidoptera: Oecophoridae) From Cambodia, Part II: five new species, checklist and taxonomic key for the species in Cambodia. J Nat Hist. 52:607–620.
- Kim S, Kaila L, Lee S. 2016. Evolution of larval life mode of Oecophoridae (Lepidoptera: Gelechioidea) inferred from molecular phylogeny. Mol Phylo Evol. 101:314–335.
- Kim S, Lee S. 2016. Taxonomic review of genus Promalactis (Lepidoptera: Oecophoridae) from Korea: description of new species with a catalog. J Asia-Pacific Ent. 19:423–437.
- Nieukerken EJ. v, Kaila L, Kitching IJ, Kristensen NP, Lees DC, Minet J, Mitter C, Mutanen M, Regier JC, Simonsen TJ, et al. 2011. Order Lepidoptera. In: Zhang, Z.-Q. (Ed.), Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Auckland, New Zealand: Magnolia Press. Zootaxa 3148:212–221.
- Tillich N, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucl Acids Res. 45:W6–W11.