Abstract
We assembled and annotated the complete mitochondrial genome of Drechslerella brochopaga. This mitogenome is a closed circular molecule of 193,195 bp in length with a GC content of 26%, including 14 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes. We analyzed the phylogenetic relationship between D. brochopaga and other 26 ascomycetes using the 14 annotated protein-coding genes of the mitochondrial genomes.
Drechslerella brochopaga (Ascomycota, Orbiliomycetes, Orbiliales, Orbiliaceae) is a nematode-trapping fungus, which captures nematodes using the specialized hyphae called constricting rings. Recently, the nuclear genomic and transcriptome sequences of Drechslerella species (D. stenobrocha) provided a new insight into the formation of constricting rings, and essential tools to reveal the transition between dual life strategies of saprophagy and predation in the kingdom Fungi (Liu et al. Citation2014). However, the genetic landscape of their mitochondria has not been studied. In this study, we report the complete mitochondrial genome of D. brochopaga, compare the essential genetic elements with those of the non-trapping species, and investigate its phylogenetic relationship with other ascomycetous species.
The mitogenome was extracted from the whole genome sequence of a pure culture of strain YMF1.03216 (collected from the State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan). We sequenced it using second-generation sequencing technology and assembled it using CLC Sequence Viewer 7.0, and the gaps of D. brochopaga (193,195 bp) mitochondrial genome were amplified by PCR. The whole mitochondrial genome was annotated automatically using the MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and GLIMMER (https://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi) based on the old mitochondrial genetic code 4. tRNAs of D. brochopaga were annotated using tRNAscan-SE (Schattner et al. Citation2005). All ORFs were searched and identified by ORFFinder.
The complete mitogenome of D. brochopaga is a closed circular molecule of 193,195bp in length with a GC content of 26%, which contains 38 genes, including 14 core mitochondrial protein-coding (PCGs) genes, 22 tRNA genes (tRNAASN gene lacked), and 2 rRNA genes. Protein-encoding genes include three ATP synthase subunits (atp6, atp8, and atp9), three cytochrome oxidase subunits (cox1, cox2, and cox3), one apocytochrome b (cob), and seven NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). The genes were encoded on the L-stand except for ATP9 gene. All the predicted genes totally possessed 30 introns, but none of the tRNAs possessed introns. All tRNA genes are encoded on the sense strand and it does not contain the three amino acids Cysteine (Cys), Proline (Pro), and Valine (Val). Especially, 12 of 14 PCGs initiated with ATG as the start codon while ATP8 began with ATT and ATP6 started with GTG, which was not found in a close relative of this species (Dactylella sp. YMF1.01838). Dactylella sp. YMF1.01838 is not able to produce any trapping devices when contacting with nematodes. Therefore, the special start codon in D. brochopaga is probably of evolutionary significance in the fungal life strategies transition.
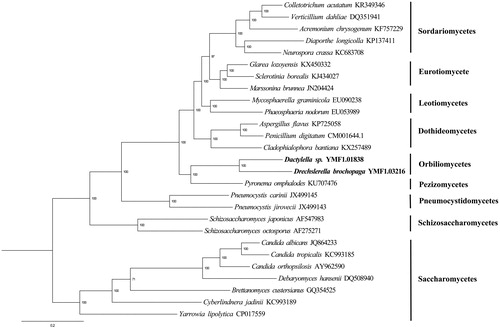
Phylogenetic analysis of 14 mitochondrial proteins of D. brochopaga and other 26 species in Ascomycota was performed by Bayesian inference (BI). As shown in , D. brochopaga and Dactylella sp. clustered together and related to Pyronema omphalodes, a member from Pezizomycetes, indicating the basal phylogenetic position of Orbiliomycetes versus most members of the Pezizomycotina. The phylogenetic relationship using mitochondrial proteins was in accordance with those constructed based on nuclear genome-scale sequences (Spatafora et al. Citation2017). The Genbank Accession number for Drechslerella brochopaga is MK550698.
Figure 1. Molecular phylogeny of 27 fungal species based on the Bayesian inference (BI) of concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The 14 mitochondrial protein-coding genes were: nad1, nad2, nad3, nad4, nad4L, nad5, nad6, cox1, cox2, cox3, cob, atp6, atp8, and atp9. The tree was generated using Bayesian inference (BI) and the best mode was LG + G + F. Numerical values along branches represent statistical support based on 1000 randomizations.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Liu KK, Zhang WW, Lai YL, Xiang MC, Wang XN, Zhang XY, Liu XZ. 2014. Drechslerella stenobrocha genome illustrates the mechanism of constricting rings and the origin of nematode predation in fungi. BMC Genomics. 15:114.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686.
- Spatafora JW, Aime MC, Grigoriev IV, Martin F, Stajich JE, Blackwell M. 2017. The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol Spectrum. 5:FUNK-0053-2016.
