Abstract
The complete chloroplast genome sequence of Sparganium eurycarpum subsp. coreanum was determined. The genome size was 161,761 bp in length with 36.9% GC content. It included a pair of inverted repeats (IRa and IRb) of 26,886 bp, which were separated by small single copy (SSC: 19,029 bp) and large single copy (LSC: 88,960 bp) regions. The cp genome contained 112 genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Phylogenetic analysis of the combined 79 protein-coding genes showed that S. eurycarpum subsp. coreanum was most closely related to Typha latifolia.
Sparganium L. comprises approximately 14–19 species and occurs in cool and temperate regions of northern and part of southern (e.g. Oceania) hemisphere (Cook and Nicholls Citation1986, Citation1987; Kaul Citation2000; Sun and Simpson Citation2010). S. erectum L. and S. eurycarpum Engelm are early diverged group among Sparganium (Sulman et al. Citation2013) and mainly distributed in Eurasia and North America, respectively. S. eurycarpum subsp. coreanum (H.Lév.) C.D.K.Cook & M.S.Nicholls has limited distribution in Korea and Japan and has been characterized by large fruits and globose rhizomes (Kadono Citation2014). However, the taxonomic status of S. eurycarpum subsp. coreanum has long been controversial (Ohwi Citation1965; Cook and Nicholls Citation1987). It is still required to determine the taxonomic status of this species and its phylogenetic position. In this study, we sequenced the complete chloroplast genome of S. eurycarpum subsp. coreanum.
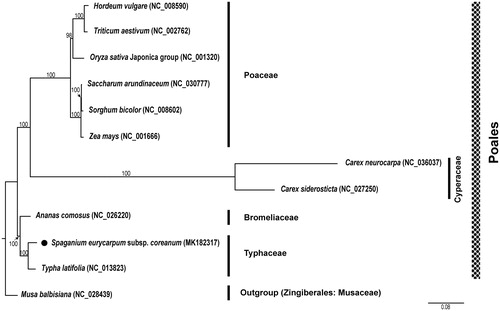
The wild individual was collected from Jeju Island, Korea (voucher specimen Gil2784; deposited in the Korea National Arboretum herbarium, KH). Total DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen Inc., Valencia, California, USA). Library preparation and genomic sequencing on the Illumina Miseq platform were conducted by the Macrogen (Macrogen Inc., Seoul, South Korea). A total of 6,592,060 raw reads were obtained and trimmed (error probability limit: 0.01) using the Geneious R v. 10.2.2 program (Biomatters Ltd., Auckland, New Zealand). The chloroplast genome was assembled with the reference chloroplast genome of Typha latifolia L., Typhaceae (Guisinger et al. Citation2010; NC_013823.1). The chloroplast genome of S. eurycarpum subsp. coreanm was annotated using Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004). The tRNA regions were confirmed using tRNAscan-SE with default setting (Schattner et al. Citation2005). Annotation of the start/stop codons of protein-coding genes was done using BLASTX, Geneious R v.10.2.2. (Biomatters Ltd., Auckland, New Zealand), and then manually corrected for intron/exon boundaries. The complete chloroplast genome of S. eurycarpum subsp. coreanum (GenBank accession MK182317) was 161,761 bp with 36.9% GC content and contained two inverted repeat regions (IRa and IRb) of 26,886 bp, separating a large single copy (LSC) region of 88,960 bp and a small single copy (SSC) region of 19,029 bp. The plastome contained 112 genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Twenty genes, including seven protein-coding genes, nine tRNA genes, and four rRNA genes, were duplicated in the IR regions. The structure, gene order, and gene content of the S. eurycarpum subsp. coreanum were similar to the Typha latifolia of Typhaceae chloroplast genomes (Guisinger et al. Citation2010). Maximum likelihood analysis of Poales was conducted based on concatenated 79 chloroplast coding genes using IQ-TREE v.1.6.8 (Nguyen et al. Citation2015) with 1000 bootstrap (BS) replications. The phylogenetic tree of Poales is shown in . The ML tree showed that Typhaceae were monophyletic (100% BS, ). S. eurycarpum subsp. coreanum was most closely related to Typha latifolia.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cook CDK, Nicholls MS. 1986. A monographic study of the genus Sparganium (Sparganiaceae). Part 1, Subgenus Xanthosparganium Holmberg. Bot Helv. 96:213–267.
- Cook CDK, Nicholls MS. 1987. A monographic study of the genus Sparganium (Sparganiaceae). Part 2, Subgenus Sparganium. Bot Helv. 97:1–44.
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 70:149–166.
- Kadono Y. 2014. A field guide to aquatic plants of Japan. Tokyo: Bunichi sogo Suhppan (in Japanese).
- Kaul RB. 2000. Sparganiaceae. In: Flora of North America Editorial Committee, editor. Flora of North America North of Mexico. Vol. 22: Magnoliophyta: Alismatidae, Arecidae, Commelinidae (in Part) and Zingiberidae. New York and Oxford: Oxford University Press; p.270–277.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ohwi J. 1965. Flora of Japan (in English). Washington, DC: Smithsonian Institution; p.119.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Sulman JD, Drew BT, Drummond C, Hayasaka E, Sytsma KJ. 2013. Systematics, biogeography, and character evolution of Sparganium (Typhaceae): Diversification of a widespread, aquatic lineage. Am J Bot. 100:2023–2039.
- Sun K, Simpson DA. 2010. Typhaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 23 Acoraceae through Cyperaceae. Beijing (China): Science Press; St, Louis (MI): Missouri Botanical Garden Press; p.158–163.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.