Abstract
Heracleum moellendorffii Hance is a traditional widely used medicinal plant in China, also known for an edible wild herb. The complete chloroplast genome of H. moellendorffii was 149,349 bp in length, composed of a large single-copy (LSC) and a small single-copy (SSC) region joined by two identical inverted repeat regions (IRA and IRB). The cp genome contained 130 genes, including 85 protein-coding genes, eight rRNA genes, and 37 tRNA genes. The GC content was 37.5%. Phylogenetic tree indicated that H. moellendorffii was close to Pastinaca pimpinellifolia.
Keywords:
Heracleum moellendorffii Hance, which subjects to the genus Heracleum L., Tordylinae in Apiaceae family (Logacheva et al. Citation2010; Yu et al. Citation2011), is a perennial shade herb widely dispersing within diverse habitants below 3200 m in China, Korea, and Japan. In its many producing areas, H. moellendorffii has long been used for not only treating cold and rheumatic arthritis, but also an edible herb. Some pharmacological studies had shown that the active ingredients of H. moellendorffii had favorable effects on anti-oxidative (Bang et al. Citation2009) and anti- melanogenesis (Alam et al. Citation2016). What is more, researchers had also made extensive attempts to apply crude extracts of H. moellendorffii to agricultural disease control (Han and Zhu Citation2009). Although H. moellendorffii has been cultivated in some areas, bulks of wild resources have been excessively collected and difficult to recover, similarly, a tough situation are forcing the whole genus Heracleum. Hence, we reported the cp genome of H. moellendorffii, which will be conducive for further research.
Mature leaves of H. moellendorffii were collected from a grassy slope near Songpan county (32°64′N, 103°61′E, altitude 2850 m), Sichuan Province, China, and the voucher specimen (voucher number:KL2018090101-1) was deposited in Sichuan University Herbarium (SZ). The extracted total DNA was then used for library building and high-throughput sequencing by Illumina Hiseq PE150 platform (Illumina, San Diego, CA), and more than 22GB raw data of paired-end reads was obtained. FastQC (v0.11.8) and Cutadapt were used to conduct quality control and clean data was used for de novo assembly by NOVOplasty2.7.2 . DOGMA (Wyman et al. Citation2004) and Geneious11.1.5 (Kearse et al. Citation2012) were used for annotating the protein-encoding genes and rRNA genes and tRNAscan (Lowe and Chan Citation2016) to authenticate the tRNA genes, all predicted genes were manually proofread. Finally, 13 complete chloroplast sequences of allied genera were aligned by MUFFT(Katoh et al. Citation2002) and trimmed for construction of maximum-likelihood (ML) tree by MEGA7.0 (Kumar et al. Citation2016) with GTR + G model,1000 bootstrap replicates was set.
The circular double-stranded complete chloroplast genome (GenBank accession no.MK210561) was 149,349 bp in length, exhibiting a quadripartite structure with a large single-copy region (LSC) of 92,199 bp and a small single-copy region (SSC) of 17,522 bp joined by two identical inverted repeat regions (IRA and IRB) of 19,814 bp. A total of 130 genes, including 85 protein-coding genes, 8 rRNA genes, and 37 tRNA genes were annotated. Seventeen genes duplicated in the inverted repeat regions. The GC content was 37.5%.
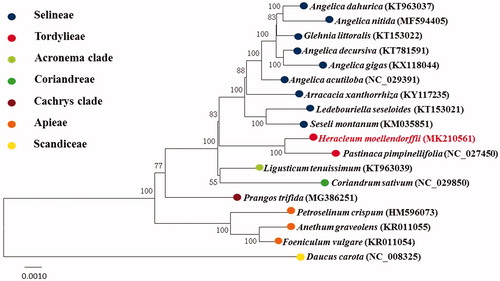
As was observed in the previous study (Downie et al. Citation2010), the phylogenetic tree () indicated that H. moellendorffii acted as a sister group to Pastinaca pimpinellifolia with 100% BS value, and Tordylinae was close to Selineae. The organelle genome can be used for phylogenetic inference and protection of wild resources.
Acknowledgment
The authors appreciate people who disclosed cp genomes data in NCBI. The authors thank Xian-Lin Guo, Fang-Yu Jin, Chuan Xie for the help of manuscript revision.
Disclosure statement
The authors declare no potential conflict of interest.
Additional information
Funding
References
- Alam MB, Seo B-J, Zhao P-J, Lee S-H. 2016. Anti-melanogenic activities of Heracleum moellendorffii via ERK1/2-mediated MITF downregulation. IJMS. 17:1844.
- Bang JE, Choi HY, Kim SI. 2009. Anti-oxidative activity and chemical composition of various Heracleum moellendorffii Hance extracts. Korean J Food Preserv. 16:765–771.
- Downie SR, Spalik K, Katz-Downie DS, Reduron JP. 2010. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Div Evol. 128:111–136.
- Han J-h, Zhu M-j. 2009. Studies on fungistasis of 27 plants. J Northwest Sci-Tech Univ Agri and for (Nat.Sci.Ed). 30 (in Chinese).
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for theorganization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–5W57.
- Logacheva MD, Valiejo-Roman CM, Degtjareva GV, Stratton JM, Downie SR, Samigullin TH, Pimenov MG, et al. 2010. A comparison of nrDNA ITS and ETS loci for phylogenetic inference in the Umbelliferae: an example from tribe Tordylieae. Mol Phylogenet Evol. 57:471–476.
- Marcel M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12.
- Dierckxsens N, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yu Y, Downie SR, He X, Deng X, Yan L. 2011. Phylogeny and biogeography of Chinese Heracleum (Apiaceae tribe Tordylieae) with comments n their fruit morphology. Plant Syst Evol. 296:179–203.