Abstract
Eriocaulaceae, a family of herbaceous plants in the order Poales, includes some well-known species used in traditional Chinese herbal medicine. The phylogenetic position of Eriocaulaceae in Poales remains controversial. In this study, we reported the complete chloroplast genome of Eriocaulon sexangulare (Eriocaulaceae) assembled from Illumina sequencing reads. The complete chloroplast genome was 151,435 bp in length with 38.60% overall GC content, containing a large single copy (LSC) region of 81,533 bp, a small single copy (SSC) region of 17,114 bp and a pair of inverted repeat (IR) regions of 26,394 bp. A total of 134 genes were annotated, including 88 protein-coding genes, 8 rRNAs, and 38 tRNAs. Phylogenetic analysis showed that E. sexangulare is most closely related to Syngonanthus chrysanthus (Eriocaulaceae) and the family Eriocaulaceae is sister to Mayacaceae.
Eriocaulaceae, a family of herbaceous plants in the order Poales, occurs in tropical and subtropical regions. Species in this family prefer plashy and aquatic environment, such as marsh, rice field, and sandy ponds (Ma et al. Citation2000). Some species in Eriocaulaceae, especially those in the genus Eriocaulon L. have been used as traditional Chinese herbal medicine for more than 1000 years (e.g. E. sexangulare L., E. buergerianume Körn., E. australe R.Br., etc.) as phenols with high antibacterial activity are enriched in their inflorescences and scapes (Li et al. Citation2009). The phylogenetic position of Eriocaulaceae in Poales remains controversial. Givinish et al. (Citation2010) proposed that Eriocaulaceae was sister to Mayacaceae and the two families clustered with Xyridaceae as the xyrid clade. However, according to APG IV, the xyrid clade comprised only Eriocaulaceae, Xyridaceae, and Mayacaceae was placed in cyperid clade (Angiosperm Phylogeny Group Citation2016). Genomic resources of Eriocaulaceae would be valuable for future phylogenetic study and utilization of this family. Therefore, we sequenced and characterized the complete chloroplast genome of E. sexangulare.
Fresh leaves of an E. sexangulare individual were collected from Maluanshan, Shenzhen, Guangdong, China. Total genomic DNA was extracted using a Plant Genomic DNA Kit (Magen, Shenzhen, China). The isolated genomic DNA was used to construct a DNA library with 350 bp insert size and sequenced on an Illumina HiSeq2500 platform (Novogene, Beijing, China). In total, 12.1 Gb 150 bp paired-end sequence data was used to assemble its complete chloroplast genome using NOVOPlasty (Dierckxsens et al. Citation2017) with the rbcL sequence of E. buergerianum (GenBank accession KC123360) as the seed. DOGMA (Wyman et al. Citation2004) was used to annotate the assembled chloroplast genome coupled with manual corrections towards start and stop codons.
The complete chloroplast genome of E. sexangulare (GenBank accession MK193813) was 151,435 bp in length with 38.60% GC content, consisting of a large single-copy (LSC) region of 81,533 bp and a small single-copy (SSC) region of 17,114 bp, separated by a pair of identical inverted repeat regions (IRs) of 26,394 bp each. A total of 134 genes were predicted, including 88 protein coding genes, 8 rRNA genes, and 38 tRNA genes.
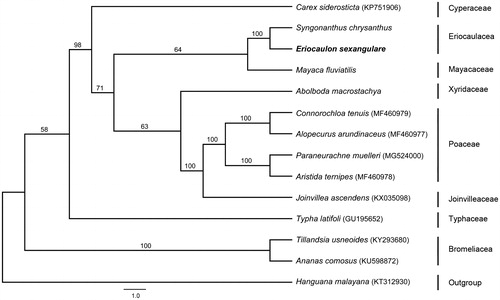
To investigate the phylogenetic relationship of Eriocaulaceae within the order Poales, four species from the famliy Poaceae, two from Bromeliaceae, and one each from Cyperaceae, Joinvilleaceae, Typhaceae, Eriocaulaceae, Mayacaceae and Xyridaceae were included in the analysis. Hanguana malayana (Commelinales) was selected as an outgroup. A combination of 30 protein-coding gene sequences shared by these 14 species were aligned using MAFFT (Katoh and Standley Citation2013). The phylogenetic relationships were inferred using maximum-likelihood method performed with RAxML (Stamatakis Citation2014). Phylogenetic analysis showed that E. sexangulare is most closely related to Syngonanthus chrysanthus (Eriocaulaceae) and the family Eriocaulaceae is sister to Mayacaceae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Givinish TJ, Ames M, McNeal JR, McKain MR, Steele PR, dePamphilis CW, Graham SW, Pires JC, Stevenson DW, Zomlefer WB, et al. 2010. Assembling the tree of the monocotyledons: plastome sequence phylogeny and evolution of Poales. Ann Mo Bot Gard. 97:584–616.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li XY, Su YG, Chen XJ, Liu W, Sun ZL. 2009. Studies on Effective Composition Analysis and in Vitro Antibacterial Activity of Eriocaulon Buergerianum Koern. Pratacult Anim Husb. 5:10–12.
- Ma WL, Zhang ZX, Stützel T. 2000. Eriocaulaceae. Vol 24. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanicl Garden Press; p. 7–17.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.