Abstract
Hermissenda emurai is a species of sea slugs in the northwestern Pacific region. In this study, the complete mitochondrial genome of H. emurai (Mollusca: Facelinidae) collected from Korea was recorded. The assembled mitogenome of H. emurai was circular and 13,880 bp in length, including 27.1% A, 15.9% C, 20.5% G, and 36.5% T nucleotide distributions. The overall A–T composition was 63.6% while the G–C composition was 36.4%. Phylogenetic analysis according to 12 protein coding genes (atp8 gene excluded) showed that H. emurai clustered with Sakuraeolis japonica in the order Nudibranchia.
Hermissenda emurai is a northwestern Pacific nudibranch in the family Facelinidae (WoRMS Editorial Board 2018). This species is found from Japan to Korea and Russian far east (Lindsay and Valdés 2016). Since the partial mitochondrial gene based study suggested the family Facelinidae is not monophyletic, the phylogenic relationship of the family is still under investigation to resolve the phylogenetic issue (Carmona et al. Citation2013). In phylogeny studies, the complete mitogenome is known to provide better phylogenetic resolution and accuracy compared to single genetic marker (Duchêne et al. Citation2011). However, until now, Sakuraeolis japonica is the only species from the family Facelinidae with the complete mitogenome available (Karagozlu et al. Citation2016). More mitogenome sequences should be recorded to resolve the phylogeny of the family. Therefore, the present study was performed to sequence and analyze the complete mitogenome of H. emurai.
The H. emurai specimen was collected in May 2013 from Gyeongpo, Korea (37°48′34.6″N 128°54′43.3″E) and deposited in Department of Biotechnology, Sangmyung University, Korea (SMU0024). Following collection, the specimen was stored in 97% ethanol for DNA extraction. The genomic DNA extracted from the specimen was used for sequencing on MiSeq system (Illumina, CA, USA). The sequence assembly and annotation were performed following the description by Karagozlu et al. (Citation2016). The phylogenetic tree was constructed based on amino acid sequences of 12 protein coding genes (atp8 gene excluded) to confirm phylogenetic relationships of H. emurai in the order Nudibranchia. The neighbor-joining method with 1000 bootstrap replicates in MEGAX software was conducted for the tree construction (Kumar et al. Citation2018). The species Ascobulla fragilis and Elysia ornata from the superorder Sacoglossa were used as an outgroup.
The mitochondrial genome of H. emurai (GenBank accession number: MK279704) was 13,880 bp in length and consists of the 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, and 22 tRNA genes. The H. emurai mitogenome had the common gene arrangement of nudibranchs (Sevigny et al. Citation2015). The overall nucleotide composition of the mitogenome was 27.1% A; 15.9% C; 20.5% G; 36.5% T and A–T accounted for 63.6% of the total nucleotide.
The typical ATG start codon was found in the eight PCGs (atp6, cox1, cox3, cytb, nd2, nd3, nd4, and nd5) while nd6 and nd4l started with ATT, cox2 and atp8 started with ATA, and nd1 started with TTG. For termination, nine PCGs (atp6, cox1, cox2, cox3, nd2, nd3, nd4l, nd5, nd6) used TAA codon for stop, three PCGs (nd1, cytb, nd4) ended with TAG and a gene (atp8) had incomplete stop codon (T–).
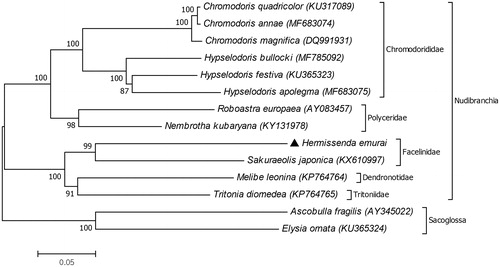
The phylogenetic tree showed that the most related species to H. emurai is Sakuraeolis japonica, which also belongs to the family Facelinidae (). The finding is congruent to previous studies on nudibranch phylogeny based on protein coding genes of mitogenomes (Sevigny et al. Citation2015; Karagozlu et al. Citation2016). Although our result provides additional data for the molecular phylogeny of the family Facelinidae, this is the second record for the family. Hence for further studies, additional data is required to understand the phylogeny of the family.
Figure 1. The phylogenetic relationships of Hermissenda emurai and related nudibranchs based on amino acid sequences of 12 mitochondrial protein coding genes (atp8 gene excluded). GenBank accession numbers of the mitogenome sequences were listed behind the species names. The species Ascobulla fragilis and Elysia ornata from the superorder Sacoglossa were used as outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Carmona L, Pola M, Gosliner TM, Cervera JL. 2013. A tale that morphology fails to tell: a molecular phylogeny of Aeolidiidae (Aeolidida, Nudibranchia, Gastropoda). PLoS One. 8:e63000.
- Duchêne S, Archer FI, Vilstrup J, Caballero S, Morin PA. 2011. Mitogenome phylogenetics: The impact of using single regions and partitioning schemes on topology, substitution rate and divergence time estimation. PLoS One. 6:e27138.
- Karagozlu MZ, Sung J-M, Lee JHyun, Kim S-G, Kim C-B. 2016. Complete mitochondrial genome analysis of Sakuraeolis japonica (Baba, 1937) (Mollusca, Gastropoda, Nudibranchia). Mitochondrial DNA B. 1:720–721.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lindsay T, Valdés Á. 2016. The model organism Hermissenda crassicornis (Gastropoda: Heterobranchia) is a species complex. PLoS One. 11:e0154265.
- Sevigny JL, Kirouac LE, Thomas WK, Ramsdell JS, Lawlor KE, Sharifi O, Grewal S, Baysdorfer C, Curr K, Naimie AA, et al. 2015. The mitochondrial genomes of the nudibranch mollusks, Melibe leonine and Tritonia diomedea, and their impact on gastropod phylogeny. PLoS One. 10:e0127519.
- WoRMS Editorial Board. 2018. World register of marine species. [Accessed 2018 December 9]. http://www.marinespecies.org at VLIZ.
