Abstract
Myoxocephalus scorpius (Linnaeus, 1758) is the second abundant fish species in coastal Arctic ecosystems with very less genetic information. In this study, the complete mitochondrial genome sequence of M. scorpius was sequenced with 16,626 bp in length, including the structure of 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and two non-coding regions (origin of light strand replication and control region). All the protein-coding genes use ATG as start codon except COI using GTG. Most of them have TAA or TAG as the stop codon while COII, ND4, and Cyt b use T–. Phylogenetic tree involving all the available Arctic fish was also reconstructed and can provide further genetic analysis for this species.
The shorthorn sculpin, Myoxocephalus scorpius (Linnaeus, 1758) belongs to the family Cottidae of suborder Cottoidei (Robins and Ray Citation1986), which is the second abundant fish species in coastal Arctic ecosystems and distribute mainly from temperate to Arctic waters (Landry et al. Citation2018). Despite its importance to Arctic ecosystems, very less is known about genetic information (Mecklenburg and Steinke, Citation2015).
The voucher specimen was collected from Chukchi Sea continental shelf (168.8°W, 69.6°N) during the 8th Chinese National Arctic Research Expedition in 2017 and deposited at the Laboratory of Marine Biology and Ecology, MNR. In this study, the complete mitochondrial genome (mitogenome) of M. scorpius was sequenced and phylogenetic topology was reconstructed.
Long PCR and primer walking methods were employed to amplify the mitogenome sequence of M. scorpius with 37 pairs of primers. It was a typical circular form with 16,626 bp in length (GenBank number MK321578) and comprised two non-coding and 37 coding regions. The 37 coding regions include 13 protein-coding genes (PCGs), 22 tRNAs genes, two rRNA genes (12S and 16S), and two mainly non-coding regions (control region/CR and light-strand replication origin/OL). The arrangement of these genes was in accordance with that found in other marine fishes mitogenome (Wang et al. Citation2018). Four nucleoside base composition (%) is 27.2 (A), 16.8 (G), 26.8 (T), and 29.2 (C), respectively. Except for one protein-coding gene ND6 and eight tRNAs genes (-Gln, -Ala, -Asn, -Cys, -Tyr, -Ser, -Glu, and -Pro), all other genes were located on the heavy strand (H-strand).
Total length of 13 protein-coding genes was 11,433 bp and they encoded 3800 amino acids. All protein-coding genes in this species had two types of start codons, including GTG for CO and ATG for others. In addition, 11 of the 13 PCGs had completed termination codon TAA or TAG while incomplete stop codon (T–) were found in COII, ND4, and Cyt b. 21 overlaps and spaces existed in adjacent genes, such as ND1 (4 bp spaces), tRNAAsp (7 bp spaces), tRNATrp (1 bp spaces), ATP8 (10 bp overlaps), and ND4L (7 bp overlaps). In addition, 2 bp overlaps were discovered within NAD3 gene, similar to the single nucleotide frameshift insertions in many birds (Mindell et al. Citation1998). The putative OL is 39 bp long, located in the WANCY region (tRNATrp-tRNAAla-tRNAAsn-tRNACys-tRNATyr). The CR of M. scorpius located between the tRNAPro and the tRNAPhe genes is determined to be 966 bp in length.
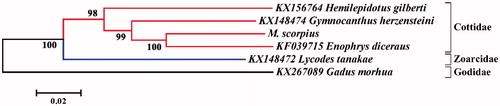
To explore the evolutionary position of M. scorpius, a neighbor-joining (NJ) tree was constructed based on the mitogenome sequences of this species and five other Arctic fish species using MEGA 6.06 (Tamura et al. Citation2013). The result showed that all species were obviously differentiated from each other with the high bootstrap values (). The taxonomic status of M. scorpius was closely related to three species of the family Cottidae, and then they clustered with other Arctic fish species. The complete mitochondrial genome and reconstructed phylogenetic relationship of M. scorpius reported in this study can provide useful genetic information for further analysis of the molecular identification, population genetics, and conservation genetics.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Landry JJ, Kessel ST, McLean MF, Ivanova SV, Hussey NE, O’Neill C, Vagle S, Dick TA, Fisk, AT. 2018. Movement types of an Arctic benthic fish, Shorthorn Sculpin (Myoxocephalus scorpius), during open water periods in response to biotic and abiotic factors. Can J Fish Aquat Sci. doi:10.1139/cjfas-2017-0389.
- Mecklenburg CW, Steinke D. 2015. Ichthyofaunal baselines in the Pacific Arctic region and RUSALCA study area. Oceanography. 28:158–189.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15:1568–1571.
- Robins CR, Ray GC. 1986. A field guide to Atlantic coast fishes of North America. Boston (MA): Houghton Mifflin Company, 354 pp.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. MEGA
- Wang SQ, Zhao LL, Li Y, Zhang ZH, Wang ZL, Gao TX. 2018. Complete mitochondrial DNA genome of grey pomfret Pampus cinereus (Bloch, 1795) (Perciformes: Stromateidae). Mitochondrial DNA B Resour. 3:683–684.