Abstract
Potentilla centigrana Maxim. is one of species in core Potentilla group. To clarify phylogenetic position of P. centigrana, we sequenced the complete chloroplast genome which is 156,392 bp long and has four subregions: 85,479 bp of large single copy (LSC) and 18,845 bp of small single copy (SSC) regions are separated by 26,034 bp of inverted repeat (IR) regions including 129 genes (84 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 37.0% and those in the LSC, SSC, and IR regions are 35.0%, 30.8%, and 42.7%, respectively. Phylogenetic trees show that P. centigrana is located far from core group of Potentilla, disagreeing with morphological classification.
Potentilla centigrana Maxim., distributed in Korea, Japan, China, and eastern Russia, presents its own morphological characters, including fibrous root, inflorescence, epicuticular waxes on both side of leaves (Heo et al. Citation2013), in comparison to other Potentilla species. It was considered one of members in Potentilla core group because phenotype of P. centigrana is similar to that of Potentilla chinensis (Wolf Citation1908); however, there is no phylogenetic study of P. centigrana till now. Moreover, there is no complete chloroplast genome of this species or neighbor species. Here, we present complete P. centigrana chloroplast genome to clarify its phylogenetic position.
Total DNA of P. centigrana collected in Republic of Korea (Voucher in InfoBoss Cyber Herbarium (IN); K-I. Heo, IB-00572) was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008), gap sequences were filled by SOAPGapCloser 1.12 (Zhao et al. Citation2011) and confirmed based on alignments generated by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Dasiphora fruticosa chloroplast complete genome (NC_036423).
The chloroplast genome of P. centigrana (MK209637) is 156,392 bp (GC ratio is 37.0%): 85,479 bp of large single copy (GC ratio is 35.0%) and 18,845 bp of small single copy (GC ratio is 30.8%) regions are separated by 26,034 bp of inverted repeats (GC ratio is 42.7%). It contained 129 genes (84 protein-coding genes, eight rRNAs, and 37 tRNAs); 17 genes (six protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in inverted repeat regions.
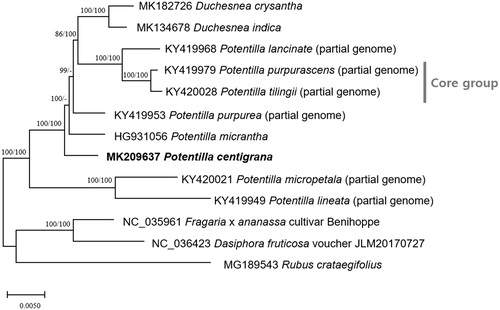
Based on currently available chloroplast genomes including six partial (Zhang et al. Citation2017) and two complete Potentilla (Ferrarini et al. Citation2013; this study), two Duchesnea, and three additional Rosaceae chloroplast genomes, neighbor joining (NJ; 10,000 bootstrap repeats) and maximum likelihood (ML; 1,000 bootstrap repeats) phylogenic trees were constructed using MEGA X (Kumar et al. Citation2018) via whole chloroplast genome sequence alignment by MAFFT 7.388 (Katoh and Standley Citation2013). Interestingly, phylogenetic trees show that P. centigrana is located outside of P. micrantha with high bootstrap support, which is far from Potentilla core group (). ML tree shows that P. micrantha and Potentilla purpurea are clustered in one clade; however, NJ tree presents that they are not (), indicating that additional species will be needed for precise phylogenetic position of P. centigrana. Moreover, there are two Potentilla species early diverged than P. centigrana: Potentilla micropetala clustered in early diverged Potentilla clade (Eriksson et al. Citation2015) and Potentilla lineata distributed in Himalaya area (Ikeda and Ohba Citation1993) indicating Himalayan clade identified in phylogenetic tree of nuclear sequences (Feng et al. Citation2017). Newly defined phylogenetic position of P. centigrana based on chloroplast genome requires researches of additional chloroplast genomes to confirm it.
Figure 1. Maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees of thirteen Rosaceae partial or complete chloroplast genomes: Potentilla centigrana (MK209637; this study), Potentilla tilingii (KY420028; partial genome), Potentilla lancinata (KY419968; partial genome), Potentilla purpurea (KY419953; partial genome), Potentila purpurascens (KY419979; partial genome), Potentilla micropetala (KY420021; partial genome) Potentilla micrantha (HG931056), Potentilla lineata (KY419949; partial genome), Duchesnea chrysantha (MK182726) Duchesnea indica (MK134678), Dasiphora fruticosa (NC 036423), Fragaria x ananassa cultivar Benihoppe (NC_035961), and Rubus crataegifolius (MG189543). The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Eriksson T, Lundberg M, Töpel M, Östensson P, Smedmark JE. 2015. Sibbaldia: a molecular phylogenetic study of a remarkably polyphyletic genus in Rosaceae. Plant Syst Evol. 301:171–184.
- Feng T, Moore MJ, Yan MH, Sun YX, Zhang HJ, Meng AP, Li XD, Jian SG, Li JQ, Wang HC. 2017. Phylogenetic study of the tribe Potentilleae (Rosaceae), with further insight into the disintegration of Sibbaldia. J Syst Evol. 55:177–191.
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, Sargent DJ. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genom. 14:670
- Heo K-I, Lee S, Yoo M, Lee S, Kwon Y, Lim SY, Kim S, Kim S-C. 2013. The taxonomic implication of trichome and epicuticular waxes in tribe Potentilleae (Rosaceae) in Korea. Korean J Plant Taxon. 43:106–117.
- Ikeda H, Ohba H. 1993. A systematic revision of Potentilla lineata and allied species (Rosaceae) in the Himalaya and adjacent regions. Bot J Linnean Soc. 112:159–186.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Wolf FT. 1908. Monographie der Gattung Potentilla. Stuttgart: E. Schweizerbartsche Verlagsbuchhandlung
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214:1355–1367.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
