Abstract
Duchesnea chrysantha (Zoll. & Moritzi) Miq. is a traditional medicinal species of genus Duchesnea nested in Potentilla genus but having a significant key to be independent genus. In this study, we presented complete chloroplast genome of D. chrysantha which is 156,387 bp long and has four subregions: 85,606 bp of large single copy (LSC) and 18,778 bp of small single copy (SSC) regions are separated by 26,002 bp of inverted repeat (IR) regions including 129 genes (84 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 37.0% and those in the LSC, SSC, and IR regions are 34.9%, 30.5%, and 42.8%, respectively. Phylogenetic trees show that two D. chrysantha are clustered and distance between D. chrysantha and D. indica is enough to be separated species.
Genus Duchesnea has its own morphological characteristic distinguished from genus Potentilla: receptacle is developed like strawberry (Chaoluan et al. Citation2003); however, it is nested in Potentilla clade (Töpel et al. Citation2011; Feng et al. Citation2017; doi:10.1080/23802359.2018.1553527, doi:10.1080/23802359.2019.1565964). Duchesnea chrysantha (Zoll. & Moritzi) Miq., one of two Duchesnea species, is distributed throughout Asia (Naruhashi and Sugimoto Citation1996). Leaflets and fruits size ranges of Duchesnea indica used for distinguishing between two species (Chaoluan et al. Citation2003) are overlapped with those of D. chrysantha (Heo et al., in submission), leading us to uncover its whole chloroplast genome to investigate phylogenetic distance between Duchesnea species.
Here, we report complete chloroplast genome of D. chrysantha collected in Korea (Voucher in InfoBoss Cyber Herbarium (IN); K Lee, IB-00578). Total DNA was extracted from fresh leaves using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea. Chloroplast genome was completed by Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011) with confirming all bases by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on previously published chloroplast complete genome of D. chrysantha (MK182726; doi:10.1080/23802359.2019.1565964).
The chloroplast genome of Korean D. chrysantha (Genbank accession is MK301251) is 156,387 bp long and has four subregions: 85,606 bp of large single copy (LSC) and 18,778 bp of small single copy (SSC) regions are separated by 26,002 bp of inverted repeat (IR) same to that of D. chrysantha chloroplast genome (MK182726; doi:10.1080/23802359.2019.1565964). It contains 129 genes (84 protein-coding genes, eight rRNAs, and 37 tRNAs); 18 genes (seven protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions. The overall GC content of Korean D. chrysantha is 37.0% and those in the LSC, SSC, and IR regions are 34.9%, 30.5%, and 42.8%, respectively.
Based on pair-wise alignment of both Duchesnea chloroplast, 48 single nucleotide polymorphisms (SNPs) and 58 insertions and deletions are identified. Interestingly, three insertion regions (6223-6242 bp, 29,455-29,472 bp, and 115,538-115,557 bp) are found on chloroplast genome of Korean D. chrysantha, contributing increase of its length by 58 bp. No variation is found in IR region. 14 of 48 SNPs (29.2%) are found inside six protein-coding genes: rpl16 has seven, ycf1 covers three, and rpoC2, cemA, rpoA, and ndhG have one SNP each.
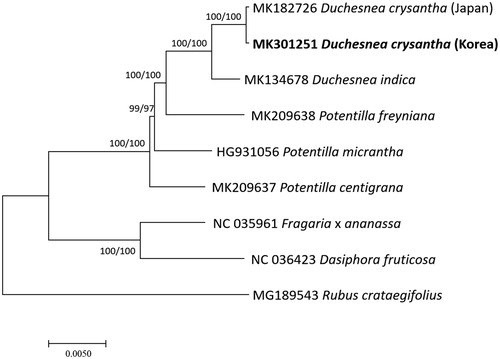
Three Duchesnea, three Potentila, and three outgroup Rosaceae complete chloroplast genome sequences were used for drawing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) trees using MAFFT 7.388 (Katoh and Standley Citation2013) and MEGA X (Kumar et al. Citation2018). Phylogenetic trees show that two D. chrysantha are clustered and distance between D. chrysantha and D. indica is enough to be considered as separated species (). This chloroplast genome will be a fundamental resource for understanding genetic diversity based on geographical distribution of D. chrysantha.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of nine Rosaceae complete chloroplast genomes: two Duchesnea chrysantha (MK301251 in this study and MK182726), Duchesnea indica (MK134678), Potentiila freyniana (MK209638), Potentilla micrantha (HG931056), Potentilla centigrana (MK209637), Dasiphora fruticosa (NC 036423), Fragaria x ananassa cultivar Benihoppe (NC_035961), and Rubus crataegifolius (MG189543). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Chaoluan L, Ikeda H, Ohba H. 2003. Potentilla Linnaeus. Sp. Pl. 1: 495. 1753. Flora of China. 9:291–327.
- Feng T, Moore MJ, Yan MH, Sun YX, Zhang HJ, Meng AP, Li XD, Jian SG, Li JQ, Wang HC. 2017. Phylogenetic study of the tribe Potentilleae (Rosaceae), with further insight into the disintegration of Sibbaldia. J Syst Evol. 55:177–191.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Naruhashi N, Sugimoto M. 1996. The floral biology of Duchesnea (Rosaceae). Plant Species Biol. 11:173–184.
- Töpel M, Lundberg M, Eriksson T, Eriksen B. 2011. Molecular data and ploidal levels indicate several putative allopolyploidization events in the genus Potentilla (Rosaceae). PLoS Curr. 3:RRN1237.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
