Abstract
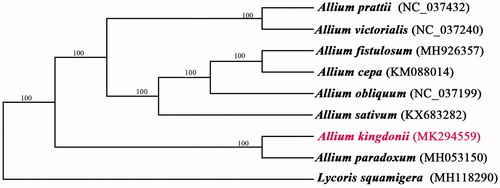
Allium kingdonii is an endemic species in southeastern Tibet, China. Here, we reported the complete chloroplast genome of A. kingdonii using the next generation sequencing method. The chloroplast genome of A. kingdonii was 153,559 bp in length which was composed of four unique regions, including a large single copy region (LSC) of 83,423 bp, a small single copy region (SSC) of 17,810 bp, and a pair of inverted repeat (IR) regions with 26,163 bp. The genome annotation predicted 132 genes, including 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Phylogenetic tree analyses suggested that A. kingdonii was closely clustered with A. paradoxum.
The Allium L. is one of the largest genera of the petaloid monocotyledons that is widely spread across the Holarctic regions from the dry subtropics to the boreal zone (Fritsch et al. Citation2010). The A. kingdonii Stearn distribute in the southeastern Tibet, it grows in hillside wetlands or shrubs with altitude ranging from 4500 m to 5000 m. Phylogenetic position of A. kingdonii has undergone a series of changes in recent years. It was always considered to have a close relationship with A. forrestii and A. changduense in the traditional classification system (Stearn Citation1960; Xu Citation1980). According to Friesen et al. (Citation2006), the A. kingdonii was belonged to Subgenus Cyathophora (R.M. Fritsch). However, a new section Kingdonia X.J. He et D.Q. Huang for A. kingdonii was proposed, and A. kingdonii was unequivocally placed into the subgenus Amerallium (Huang et al. Citation2014). In this study, we reported the complete chloroplast genome sequence of A. kingdonii, which will provide a genomic resource and can be used to investigate the phylogenetic relationships in the Allium L. with other previously reported species.
The genomic DNA was extracted following the modified CTAB method from the dry and healthy leaves (Doyle and Doyle Citation1987), the voucher specimen was collected at Nyingchi (29°37′27.18″N, 94°39′2.22″E), Tibet, China, and stored in SZ (Sichuan University Herbarium). The isolated genomic was manufactured to average 350 bp paired-end(PE) library using Illumina Hiseq platform (Illumina, San Diego, CA) and sequenced using Illumina genome platform (HiseqPE150). The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with the complete chloroplast genome of its close relative A. cepa as the reference (GenBank accession no. KM088014). The assembled chloroplast genome was annotated using Geneious 11.0.4 and corrected manually (Kearse et al. Citation2012). The complete chloroplast genome of nine species was aligned using MAFFT (Katoh et al. Citation2002). Nine species of Amaryllidaceae were employed to build the maximum-likelihood (ML) tree using RaxML (Stamatakis Citation2006) with 1000 bootstrap replicates was calculated.
The chloroplast genome of A. kingdonii was 153,559 bp in length (GenBank accession no.MK294559), containing a large single copy region (LSC) of 83,423 bp, a small single copy region (SSC) of 17,810 bp, and a pair of inverted repeat (IR) regions of 26,163 bp. Genome annotation predicted 132 genes, including 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The overall GC-content of the whole plastome was 36.9%, with the corresponding values in the LSC, SSC, and IR regions were34.8%, 29.9%, and 42.7%, respectively.
Phylogenetic analysis suggested that A. kingdonii is closely clustered with A. paradoxum (), which was consistent with a previous study (Huang et al. Citation2014). This complete cp genome can provide a genomic resource and can be further used for constructing phylogenetic relationships among the genus Allium L.
Acknowledgments
The authors thank the opened cp genome data in NCBI. The authors appreciate Yan Yu, Lu Kang and Hao Li for the help of sequence analysis.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Friesen N, Fritsch RM, Blattner FR. 2006. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. J Syst Evol Bot. 22:372–395.
- Fritsch RM, Blattner FR, Gurushidze M. 2010. New classification of Allium L. Subg Melanocrommyum (Webb & Berthel) Rouy (Alliaceae) based on molecular and morphological characters. Phyton. 49:145–220.
- Huang DQ, Yang JT, Zhou CJ, Zhou SD, He XJ. 2014. Phylogenetic reappraisal of Allium subgenus Cyathophora (Amaryllidaceae) and related taxa, with a proposal of two new sections. J Plant Res. 127:275–286.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Stearn WT. 1960. Allium and Milula in the Central and Eastern Nepal Himalaya. Bull Br Mus Nat Hist (Bot). 2:159–191.
- Xu JM. 1980. Allium L. Flora Reipublicae Popularis Sinicae. 14:202–213.