Abstract
We first determined the complete mitochondrial genome of Fusinus longicaudus (Gastropoda: Fasciolariidae). The circular mitochondrial genome is 16,319 bp in length, consisting of 13 protein-coding genes, 22 tRNA genes, and two ribosomal RNA genes. The nucleotide composition is 14.7% of C, 15.9% of G, 38.9% of T, and 30.5% of A. The control region with the length of 1029 bp is between tRNAPhe and COX3 genes. In 13 protein-coding genes, the nad4 gene starts with ATT, the other eleven genes start with ATG. For the stop codon, atp6, nad4l, nad5 and nad6 end with TAG, other 9 genes end with TAA. The phylogenetic tree was constructed based on 13 protein-coding genes of F. longicaudus and other 13 Neogastropoda species, Perna viridis as outgroup using the Neighbor-joining method, the result showed that F. longicaudus is most closely related to the Buccinidae. The mitochondrial genome would be helpful for the study of population genetic and phylogenetic analysis of the family Fasciolariidae.
Fusinus longicaudus (Lamarck, 1816) is widely distributed in the northwest Pacific Ocean. It inhabits in the rock area of the intertidal zone in the coastline from Korea to Vietnam. The sell is large and strong. It is a carnivorous conch (Min et al. Citation2004). Information on genetic of F. longicaudus is limited. There are some studies focused on the taxonomy of the Neogastropoda based on the gene sequences, which identify the different of F. longicaudus and others (Cunha et al. Citation2009). None study reported the genome information of any species in the family Fasciolariidae. The mitochondrial genome of F. longicaudus is still unknown.
This is the first study that determined the complete mitochondrial genome of F. longicaudus. The specimen of F. longicaudus was collected in Zhoushan, Zhejiang province, China (122.2°E, 30.3°N) and identified by morphology and deposited in Zhejiang Ocean University. The total DNA extraction was utilized the salting-out method (Aljanabi and Martinez Citation1997) with the muscle. Total genomic DNA was diluted to a final concentration of 70–80 ng/μl in 1 × TE buffer and stored at 4 °C for further analysis. The genomic DNA was prepared in 450 bp paired-end libraries. The Illumina HiSeq X Ten platform was used to perform the high-throughput sequence.
The complete mitochondrial genome of F. longicaudus is 16,319 bp in length (GenBank accession number: MK335921). The complete mitochondrial genome includes two ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and 13 protein-coding genes. The control region with the length of 1029 bp is between tRNAPhe and COX3 genes. The nucleotide composition for F. longicaudus is 14.7% of C, 15.9% of G, 38.9% of T, and 30.5% of A. In 13 protein-coding genes, the nad4 gene starts with ATT, the other eleven genes are with ATG. For the stop codon, atp6, nad4l, nad5, and nad6 end with TAG, other nine genes end with TAA. The 12S rRNA is 892 bp between the tRNAGlu and tRNAVal, and the 16S rRNA is 1397 bp between the tRNAVal and tRNALeu1.
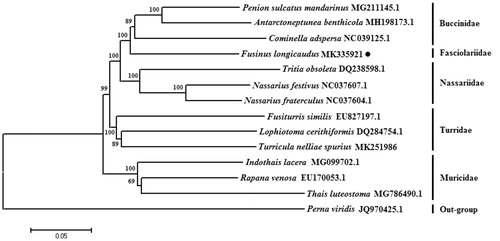
The phylogenetic tree was constructed based on 13 protein-coding genes of the F. longicaudus and other 13 Neogastropoda species, Perna viridis as outgroup using the Neighbor-joining method (Saitou and Nei Citation1987) by the program Phylip (Felsenstein Citation1989). The tree showed that the F. longicaudus is most closely related to the Buccinidae and in different branch with Muricidae, Turridae, and Nassariidae families (). We believe that this result will further supplement the genome information in mitochondria of the family Fasciolariidae and facilitate the study on population genetic.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucl Acids Res. 25:4692–4693.
- Cunha RL, Grande C, Zardoya R. 2009. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol Biol. 9:210.
- Felsenstein J. 1989. PHYLIP-Phylogeny inference package (Version 3.2). Cladistics. 5:164–166.
- Min D, Lee J, Je JJK. 2004. Mollusks in Korea. Min Molluscan Research Institute, Busan. 566.
- Saitou N, Nei M. 1987. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.