Abstract
Here, we report the first complete mitochondrial genome of a sex-limited marking silkworm (Bombyx mori), which has been exclusively developed for male-only rearing in sericulture industry. The complete mitogenome sequence is 15,678 bp in length, containing 13 protein-coding genes, 22 tRNA genes, two rRNA, and one control region. The work will provide a valuable data for future research on sex-limited inheritance and sex control in silkworm.
In sericulture, the raw silk is generally produced from first generation hybrids of silkworm, Bombyx mori. To produce silkworm hybrids, sex discrimination of parent breeds is essential. However, sex separation at the larval or pupal stages is extremely laborious and unrealistic on a commercial scale without sex-limited markings. A sex-limited marking silkworm variety, known as B. mori Xian7, has been exclusively developed for the male silkworm hybrid combination Xian7 x Ping48 (Liu et al. Citation2012). For a better understanding of its origin and breeding potential, we determined the mitochondrial genome of B. mori Xian7 for the first time.
Bombyx mori Xian7 was collected from Hangzhou City, Zhejiang Province in China (30°18′ N, 120°11′ E). The specimens have been conserved in the Silkworm Germplasm Resources Repository at Sericulture Research Institute, Zhejiang Academy of Agricultural Sciences. Total genomic DNA was extracted from a single individual using a DNeasy Tissue Kit (Qiagen). The whole mitochondrial genome was amplified using 25 pairs of primes and PCR products were determined by Sanger sequencing. DNA sequence was then analyzed using MITOS (Bernt et al. Citation2013) and tRNAscan-SE server (Schattner et al. Citation2005). The total length of the complete mitogenome is 15,678 bp (GenBank accession no. MK295814). It contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one non-coding control region. The overall base composition was 43.07% A, 38.32% T, 11.31% C, and 7.30% G, with a high A + T content of 81.39%. All PCGs start with a typical ATN codon, except COX1 gene which begins with CGA; two PCGs (COX1 and COX2) stop with incomplete codon T, whereas 11 PCGs end with usual codon TAA. The gene arrangement and orientation are identical with other lepidopteran species (Li et al. Citation2016; Zhang et al. Citation2016).
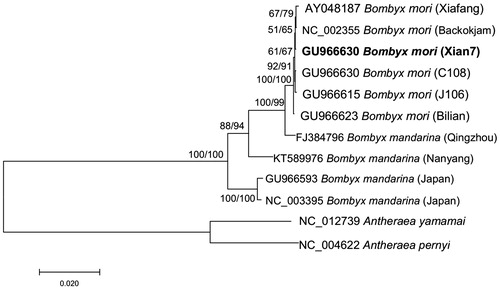
To validate the phylogenetic position of B. mori Xian7, we used MEGA 7.0 (Kumar et al. Citation2016) to construct phylogenetic trees based on the complete mitochondrial DNA sequence. Bootstrap analysis was performed with 1000 replications. The maximum-likelihood (ML) tree shows the same topological structure as the neighbor-joining (NJ) tree (). Phylogenetic analysis confirmed that B. mori Xian7 has a close genetic relationship with other silkworm varieties. The newly determined mitogenome will help to study on sex-limited inheritance and sex control in silkworm.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li FB, Zhang HX, Liu PG, Wang YQ, Meng ZQ. 2016. Complete mitochondrial genome of a hybrid strain of the domesticated silkworm (Qiufeng × Baiyu). Mitochondr DNA Part A. 27:1844–1845.
- Liu XJ, Zhu XR, Meng ZQ, Chen S, Cao JR, Ye AH, He KR. 2012. Breeding of a male silkworm variety Xian 7 × Ping 48 for summer and autumn rearing. Sci Seric. 38:0575–0580.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686.
- Zhang HX, Li FB, Zhu XR, Meng ZQ. 2016. The complete mitochondrial genome of Bombyx mori strain Baiyun (Lepidoptera: Bombycidae). Mitochondr DNA Part A. 27:1652–1653.