Abstract
In this paper, we first reported the complete mitogenomes of Psittacula derbiana and Psittacula eupatria. Respectively, P. derbiana and P. eupatria mitogenomes are 16,887 and 17,139 bp base pairs long. The overall base compositions of complete mitogenomes of P. derbiana and P. eupatria are 30.9% A, 22.4% T, 32.8% C, 13.9% G and 31.7% A, 22.0% T, 33.2% C, 13.1% G, respectively. All genes exhibit the typical mitochondrial gene arrangement and transcribing directions. The Neighbor-joining analysis suggested that Psittacula species are closely related to Eclectus roratus. The results obtained here could contribute to future studies on molecular evolution and population genetics in Psittacula species.
The genus Psittacula of the Psittacidae family includes 13 extant species in the world (Groombridge et al. Citation2004; IUCN Citation2018). Because of habitat loss and trapping pressure, most species in this genus are suspected to be undergoing a moderately rapid population decline (Olah et al. Citation2016; IUCN Citation2018). There is less information on the complete mitogenomes of the genus Psittacula. In this study, we characterized and compared the complete mitogenomes of two Psittacula species.
Whole blood samples of Psittacula derbiana and Psittacula eupatria were collected from the Nanjing Hongshan Forest Zoo (N32°09′, E118°80′) in Nanjing, Jiangsu province, China. DNA extraction was performed using the DNAiso reagent (Takara). A set of universal primers were designed for polymerase chain reaction amplification and Sanger sequencing based on the mitogenomes of Tanygnathus lucionensis (GenBank accession: KM611480.1) and Eclectus roratus (GenBank accession: KM611469.1).
The lengths of complete mitogenomes of P. derbiana and P. eupatria are 16,887 (GenBank accession: MK343133) and 17,139 bp (GenBank accession: MK343134), respectively. The two complete mitogenomes are A + T rich with the following nucleotide compositions: 30.9% A, 22.4% T, 32.8% C, 13.9% G for P. derbiana, and 31.7% A, 22.0% T, 33.2% C, 13.1% G for P. eupatria. Both mitogenomes consist of 22 transfer RNA genes, 13 protein-coding genes, two ribosomal RNA genes, and a non-coding control region. All genes follow the typical mitochondrial gene arrangement and transcribing directions, which is identical to most avian species (Boore Citation1999; Eberhard and Wright Citation2016; Caparroz et al. Citation2018).
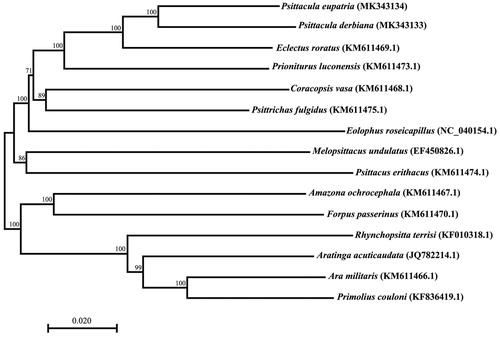
Phylogenetic analysis was performed based on the complete mitogenomes of 15 parrot species. Sequence dataset was aligned by Clustal X and analyzed using the Neighbor-joining method with 1000 bootstrap replicates in MEGA version 7.0 (Larkin et al. Citation2007; Kumar et al. Citation2016). Phylogenetic tree showed that the two Psittacula species are closely related to E. roratus (). The genome information obtained here could contribute to future studies on molecular evolution and population genetics in Psittacula species (Groombridge et al. Citation2004).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Caparroz R, Rocha AV, Cabanne GS, Tubaro P, Aleixo A, Lemmon EM, Lemmon AR. 2018. Mitogenomes of two neotropical bird species and the multiple independent origin of mitochondrial gene orders in Passeriformes. Mol Biol Rep. 2018. 45:279–285.
- Eberhard JR, Wright TF. 2016. Rearrangement and evolution of mitochondrial genomes in parrots. Mol Phylogenet Evol. 94:34–46.
- Groombridge JJ, Jones CG, Nichols RA, Carlton M, Bruford MW. 2004. Molecular phylogeny and morphological change in the Psittacula parakeets. Mol Phylogenet Evol. 31:96–108.
- International Union for Conservation of Nature (IUCN). 2018. The IUCN red list of threatened species. Psittacula. Brussels (Belgium): IUCN; Version 3.1, [accessed 2018 Oct 22]. http://www.iucnredlist.org.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948.
- Olah G, Butchart SHM, Symes A, Medina Guzmán I, Cunningham R, Brightsmith DJ, Heinsohn R. 2016. Ecological and socio-economic factors affecting extinction risk in parrots. Biodivers Conserv. 25:205–223.