Abstract
The complete mitochondrial genome of Dabieshan black pig (Sus scrofa) was first determined in this study. The mitogenome is 16,690 bp in length, consisting of 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs, and a control region. In addition, the reconstructed phylogenetic tree indicated that different Chinese pig breeds do not form a monophyletic group, and the tree also confirmed that there is certainly a genetic link between Dabieshan black pig distributed in Southwest Anhui and Huai pig distributed in North Anhui.
Dabieshan black pig (Sus scrofa) is a high-quality meat producer maybe because of varied microclimates at Dabieshan Mountain Area, mainly distributed in the southwest of Anhui Province and listed as one of the national geographical indication products in China. In this study, we present the complete mitochondrial genome of Dabieshan black pig (GenBank accession no: KP294522). The specimen was collected from a breeding pig farm at Yuexi County of Anhui in China (Altitude 850 meter, East Longitude 116.36°, Northern Latitude 30.84°). The specimen (tissue and DNA) was deposited in Provincial Key Laboratory of the Conservation and Exploitation of Important Biological Resources in Anhui (Voucher number: DBSBP-00021). Total genomic DNA was extracted from muscle tissue using the kit of genomic DNA extraction (Sangon Biotech, Shanghai, China) (Pan et al. Citation2004). Sixteen DNA fragments were amplified with 16 pairs of specific primers which were designed based on some mitochondrial DNA sequences of pigs (Yang et al. Citation2003).
The complete sequence of mitochondrial DNA of Dabieshan black pig is 16,690 bp in length. The overall nucleotide composition is 34.7% A, 25.8% T, 26.2% C, and 13.3% G, with a total A + T content of 60.5%, that is heavily biased toward A and T nucleotides. It presents the typical set of 37 genes, including 13 PCGs (COX1-3, Cyt b, ND1-6, ND4L, ATP6, and ATP8), 22 tRNA genes, two genes for ribosomal RNA subunits (12S rRNA, 16S rRNA), and a major non-coding control region (D-Loop region). ND4L gene begins with GTG as start codon, ND2, ND3, and ND5 genes begin with ATA as start codon, and other nine protein-coding genes start with ATG. Cyt b gene is terminated with AGA as stop codon, ND1 and ND2 genes are terminated with TAG as stop codon, COII, COIII, ND3, and ND4 end with T, while ATP6, ATP8, COI, ND4L, ND5, and ND6 end with TAA. In addition, the noncoding regions include a 1254-bp control region (D-loop), eight overlaps and 11 spaces in the length of 1–32 bp.
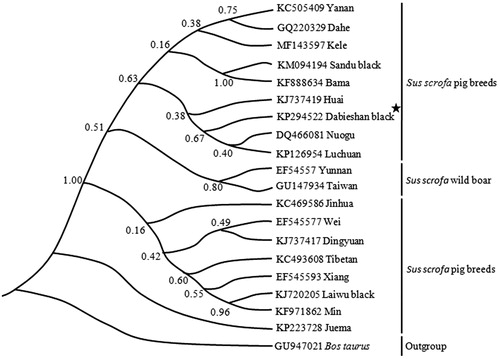
The mtDNA sequences of Dabieshan black pig, other 16 Chinese pig breeds, and two wildboar populations were used to reconstruct phylogenetic tree using MrBayes 3.2 (Ronquist et al. Citation2012), and Bos taurus was set as outgroup. The BI (Bayesian inference) phylogenetic tree () indicated that two wildboar populations form a monophyletic group, whereas 17 Chinese pig breeds do not form a monophyletic group, and Juema pig is located near the root of the molecular phylogenetic tree. Furthermore, the BI tree also confirmed that there is certainly a genetic link between Dabieshan black pig distributed in Southwest Anhui and Huai pig distributed in North Anhui.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Pan HC, Zhou KY, Song DX, Qiu Y. 2004. Phylogenetic placement of the spider genus Nephila (Araneae: Araneoidea) inferred from rRNA and MaSp1 gene sequences. Zool Sci. 21:343–351.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Yang J, Wang J, Kijas J, Liu B, Han H, Yu M, Yang H, Zhao S, Li K. 2003. Genetic diversity present within the near-complete mtDNA genome of 17 breeds of indigenous Chinese pigs. J Hered. 94:381–385.