Abstract
The mitochondrial genome (mitogenome) provides important information for phylogenetic analysis and understanding evolutionary origins. Herein, we sequenced, annotated, and characterized the mitogenome of Metaplax longipes to better understand its molecular evolution and phylogeny. The 16,424 bp mitogenome includes 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one control region. The length of control region is 1036 bp. The nucleotide composition of M. longipes is A + T: 71.7%, with a strong AT bias. Phylogenetic analysis shows that M. longipes is closely related to Cyclograpsus granulosus and Helice wuana, which belong to the same family Varunidae.
Keywords:
Metaplax longipes is a mudflat crab inhabiting Chinese coastal regions including Guangdong, Fujian, and Zhejiang and it burrows in intertidal mudflats, swamps, salt marshes, and estuaries, especially in the high intertidal and supralittoral zones. This species historically assigned to the Sesarminae, belonging to Grapsidae (Dai and Yang Citation1991). The early molecular phylogenetic analysis based on partial sequences of 12S rRNA and/or 16S rRNA revealed that Metaplax genus were included in the Varuninae and excluded from the Sesarminae (Schubart et al. Citation2000, Citation2002, Citation2006; Kitaura et al. Citation2002).
The sample of M. longipes was collected in Fujian Province. Mitogenomic DNA of M. longipes was extracted from muscle tissue sample using an Aidlab Genomic DNA Extraction Kit (Aidlab Co., Beijing, China) and stored at −20 °C until needed for PCR amplification. For amplification of the whole mitogenome of M. longipes, a set of universal primers were used to amplify a range of segments of the entire mitogenome (Tang et al. Citation2003, Citation2017, Citation2018; Simon et al. Citation2006; Xin et al. Citation2017). Subsequently, specific primers were designed using Primer Premier 5.0 and synthesized by Beijing Sunbiotech. All PCRs were performed and products were separated and purified. Purified PCR products were ligated into the T-vector (Aidlab Co., Beijing, China) and sequenced by Beijing Sunbiotech. Mitogenome sequences were searched using the BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the DNASTAR package (DNAstar, Inc., Madison, WI, USA) (Burland Citation2000). Thirteen PCGs were initially identified using an open reading frame (ORF) Finder to specify vertebrate mitochondrial genetic code and translate it into putative proteins in GenBank (Rombel et al. Citation2002) and tRNA genes were verified using MITOS WebServer (http://mitos2.bioinf.unileipzig.de/index.py) using the default setting (Bernt et al. Citation2013).
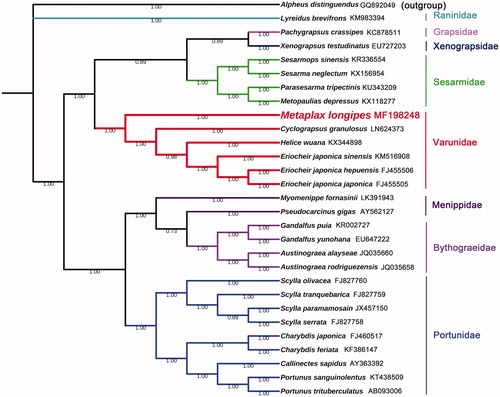
The complete mitogenome of M. longipes was determined and submitted to GenBank under accession number MF198248. The sequence was found to be 16,424 bp in size contain 13 PCGs, 22 tRNAs, two RNAs, and one control region. The nucleotide composition of the complete mitogenome was as follows A = 6154 (37.5%), T = 5612 (34.2%), G = 1709 (10.4%), and C = 2949 (17.9). The AT content was 71.7%, with a strong AT bias. For phylogenetic analysis, 29 mitogenome datasets were used to reconstruct phylogenetic relationships among Brachyura. Concatenated sets of amino acid sequences were used for phylogenetic analysis by Bayesian Inference (BI) methods using MrBayes v3.2.1 (Ronquist et al. Citation2012). The resultant phylogenetic tree indicated that M. longipes, Cyclograpsus granulosus, Helice wuana, Eriocheir japonica sinensis, Eriocheir japonica hepuensis, and Eriocheir japonica japonica were clustered in one branch with high nodal support value which is located in same family Varunidae (). Based on these results, mitogenome of M. longipes could contribute to the phylogenetic knowledge of the family Varunidae.
Figure 1. Phylogenetic tree based on 29 whole mitogenomes constructed using Bayesian Inference (BI) methods. Species used in this article: Helice wuana (KX344898), Cyclograpsus granulosus (LN624373), Eriocheir japonica sinensis (KM516908), Eriocheir japonica hepuensis (FJ455506), Eriocheir japonica japonica (FJ455505), Sesarmops sinensis (KR336554), Sesarma neglectum (KX156954), Parasesarma tripectinis (KU343209), Metopaulias depressus (KX118277), Pachygrapsus crassipes (KC878511), Xenograpsus testudinatus (EU727203), Pseudocarcinus gigas (AY562127), Myomenippe fornasinii (LK391943), Callinectes sapidus (AY363392), Portunus trituberculatus (AB093006), Portunus sanguinolentus (KT438509), Charybdis japonica (FJ460517), Scylla paramamosain (JX457150), Scylla olivacea (FJ827760), Scylla tranquebarica (FJ827759), Scylla serrata (FJ827758), Charybdis feriata (KF386147), Lyreidus brevifrons (KM983394), Gandalfus yunohana (EU647222), Gandalfus puia (KR002727), Austinograea alayseae (JQ035660), Austinograea rodriguezensis (JQ035658), and Metaplax longipes (MF198248). In addition, Alpheus distinguendus (GQ892049) used as outgroup.
Disclosure statement
No potential conflict of interest was reported by authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Burland TG. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Dai AY, Yang SL. 1991. Crabs of the China seas. Beijing: China Ocean Press.
- Kitaura J, Wada K, Nishida M. 2002. Molecular phylogeny of grapsoid and ocypodoid crabs with special reference to the genera Metaplax and Macrophthalmus. J Crustacean Biol. 22:682–693.
- Rombel IT, Sykes KF, Rayner S, Johnston SA. 2002. ORF-FINDER: a vector for high-throughput gene identification. Gene. 282:33–41.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schubart CD, Cannicci S, Vannini M, Fratini S. 2006. Molecular phylogeny of grapsoid crabs (Decapoda, Brachyura) and allies based on two mitochondrial genes and a proposal for refraining from current superfamily classification. J Zool Syst. 44:193–199.
- Schubart CD, Cuesta JA, Diesel R, Felder DL. 2000. Molecular phylogeny, taxonomy, and evolution of nonmarine lineages within the American grapsoid crabs (Crustacea: Brachyura). Mol Phylogenet Evol. 15:179–190.
- Schubart CD, Cuesta JA, Felder DL. 2002. Glyptograpsidae, a new brachyuran family from Central America: larval and adult morphology, and a molecular phylogeny of the Grapsoidea. J Crustacean Biol. 2:28–44.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37:545–579.
- Tang BP, Liu Y, Xin ZZ, Zhang DZ, Wang ZF, Zhu XY, Wang Y, Zhang HB, Zhou CL, Chai XY, Liu QN. 2018. Characterisation of the complete mitochondrial genome of Helice wuana (Grapsoidea: Varunidae) and comparison with other Brachyuran crabs. Genomics. 110:221–230.
- Tang BP, Xin ZZ, Liu Y, Zhang DZ, Wang ZF, Zhang HB, Chai XY, Zhou CL, Liu QN. 2017. The complete mitochondrial genome of Sesarmops sinensis reveals gene rearrangements and phylogenetic relationships in Brachyura. PLoS One. 12:e0179800
- Tang BP, Zhou KY, Song DY, Yang G, Dai AY. 2003. Molecular systematics of the Asian mitten crabs, genus Eriocheir (Crustacea: Brachyura). Mol Phylogenet Evol. 29:309–316.
- Xin ZZ, Yu-Liu Zhang DZ, Wang ZF, Zhang HB, Tang BP, Zhou CL, Chai XY, Liu QN. 2017. Mitochondrial genome of Helice tientsinensis (Brachyura: Grapsoidea: Varunidae): gene rearrangements and higher-level phylogeny of the Brachyura. Gene. 627:307–314.
