Abstract
The complete mitochondrial genome sequences of Heterakis dispar were determined for the first time in present study. The lengths of mtDNA sequences of H. dispar is 13,995 bp in size, containing 12 PCGs (cox1-3, nad1-6, nad4L, atp6, and cytb), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two non-coding regions, all genes are encoded by the same strand. The phylogenetic analysis by Bayesian inference method revealed that Ascaridiidae was closely related with Heterakidae than to other Ascaridida nematodes. In Heterakidae, H. dispar was closely related with H. gallinarum than to H. beramporia with high statistical support. The mtDNA dataset provided useful genetic markers for studying the molecular ecology, systematics, and population genetics of Ascaridida nematodes.
Genus Heterakis is one of the major parasitic nematodes of poultry, which has a wide geographic distribution. Heterakis dispar is one of the important member of genus Heterakis, which colonize in the caecum of chickens, geese, partridges and wild waterfowls (Permin et al. Citation1997; Avcioglu et al. Citation2008; Shutler et al. Citation2012). The parasites can cause digestive function dysfunction, anemia, emaciation, and even death, leading to considerable economic losses to the poultry industries.
However, up to date, only rare dataset reported about the parasite, particular in genetic information. In this study, we determined the complete mitochondrial genome of H. dispar isolated from caecum of a goose in Heilongjiang Province, China. The individuals were identified according to its morphological characteristic (Avcioglu et al. Citation2008) and stored in the Department of Parasitology, Heilongjiang institute of Veterinary Science (specimen no. CDL-180512). Total genomic DNA was extracted from a single individual using TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) and stored at −20 °C until use. Primers were designed for PCR amplification and sequencing base on the mt sequence of H. gallinae (GenBank No. KU529973) (Wang et al. Citation2016).
The mt genomes of H. dispar is circular molecule with 13,995 bp (GenBank no. MK024389) in size, which containing 12 PCGs (cox1-3, nad1-6, nad4L, atp6, and cytb), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two non-coding regions (NCR), all genes are encoded by the same strand. The nucleotide contents of the mt genome sequence were biased toward A + T (69.8%). The H. dispar mt genome encoded 3416 amino acids in total. The A + T contents of PCGs ranged from 66.0% (nad1) to 75.2% (nad4L). Codon usage analysis of H. dispar showed that four kinds of initiation codons (ATG, ATA, ATT, and TTG) and three kinds of termination codons (TAA, TAG, and T) were used. The size of 22 tRNAs ranging from 52 to 62 bp. The rrnL (951 bp) is located between trnH and nad3, whereas rrnS (702 bp) is located between trnE and trnS(UCN). The Long NCR (630 bp) is located between trnC and trnN, which A + T contents are 68.9%, The Short NCR (136 bp) is located between nad4 and trnM, which A + T contents are 70.6%.
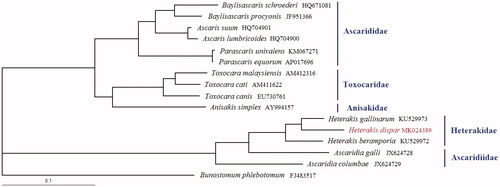
Phylogenetic analysis of H. dispar and other 14 Ascaridida nematodes were conducted based on the concatenated amino acid sequences of 12 PCGs with Bunostomum phlebotomum as outgroup using Bayesian inference (BI) methods (). Phylogenetic analysis revealed that Ascaridiidae was closely related with Heterakidae than to other Ascaridida nematodes; the result is consistent with that based on cox1 sequence previously reported (Šnábel et al. Citation2014). In Heterakidae, H. dispar was closely related with H. gallinarum than to H. beramporia with high statistical support. The availability of the complete mt genomes provided useful genetic markers for studying the molecular ecology, systematics, and population genetics of Ascaridida nematodes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Avcioglu H, Burgu A, Bölükbaş CS. 2008. Ascaridia numidae (Leiper, 1908; Travassos, 1913) in Rock Partridge (Alectoris chukar) in Turkey. Parasitol Res. 102:527–530.
- Permin A, Magwisha H, Kassuku AA, Nansen P, Bisgaard M, Frandsen F, Gibbons L. 1997. A cross-sectional study of helminths in rural scavenging poultry in Tanzania in relation to season and climate. J Helminthol. 71:233–240.
- Shutler D, Alisauskas RT, Daniel McLaughlin J. 2012. Associations between body composition and helminths of lesser snow geese during winter and spring migration. Int J Parasitol. 42:755–760.
- Šnábel V, Utsuki D, Kato T, Sunaga F, Ooi HK, Gambetta B, Taira K. 2014. Molecular identification of Heterakis spumosa obtained from brown rats (Rattus norvegicus) in Japan and its infectivity in experimental mice. Parasitol Res. 113:3449–3455.
- Wang BJ, Gu XB, Yang GY, Wang T, Lai WM, Zhong ZJ, Liu GH. 2016. Mitochondrial genomes of Heterakis gallinae and Heterakis beramporia support that they belong to the infraorder Ascaridomorpha. Infect Genet Evol. 40:228–235.