Abstract
Pseudoxenodon is a genus of snakes belonging to the family Colubridae. In this study, the mitochondrial genome sequence of Pseudoxenodon bambusicola was generated through the next-generation sequencing technique. A bayesian inference (BI) phylogenetic tree based on the 12 protein-coding genes was re-constructed for convincing the mitochondrial DNA sequences.
Pseudoxenodon, with an important diagnostic feature that is the obliquely arranged scales on the anterior part of the body, are widely distributed throughout southern and southeastern Asia (Zhang and Huang Citation2013). In this study, the complete mitochondrial genome of Pseudoxenodon bambusicola was isolated and characterized.
Specimens of P. bambusicola were collected from Quanzhou, Fujian province, China, and deposited at CIB herpetological museum, Chengdu Institute of Biology with the number CIB010247. The genomic data were generated directly through the next-generation sequencing (Hahn et al. Citation2013). The mitochondrial genome of Thermophis zhaoermii (GenBank accession number: GQ166168.1), which was downloaded from GenBank was used as a reference sequence for assembling using MITObim (Paszkiewicz and Studholme Citation2010). The annotations were generated from Mitochondrial genome annotation WebServer (MITOS) (Bernt et al. Citation2013) with manual correction and tRNAScan-SE (Lowe and Eddy Citation1997) for tRNA genes.
For phylogenetic analyses of Pseudoxenodon bambusicola, MrBayes (Version 3.2.6) (Ronquist et al. Citation2012) was used to reconstruct a bayesian inference (BI) tree with 12 H-stand protein-coding genes (PCGs) from 11 mitochondrial genomes of snakes, while the ND6 gene, which is encoded by the L-strand, was excluded. Xenopeltis unicolor (GenBank: NC_007402) was selected as an outgroup. The alignment was performed by MEGA7 (Kumar et al. Citation2016) with default settings. All gaps and poorly aligned positions were removed using Gblocks (Version 0.91b) (Castresana Citation2000). DAMBE (Xia Citation2018) was used for testing substitution saturation. The best fitting model of sequence evolution (GTR + I + G) was obtained using Modeltest 3.7 (Posada and Crandall Citation1998) on the basis of the Akaike information criterion (AIC) (Akaike Citation1974).
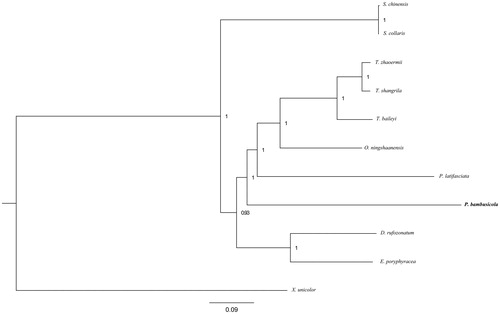
As a result, the mitogenome sequence of P. bambusicola (GenBank accession number: MK238278) consists of 17,725 bp length and contains 2 ribosomal RNA genes (12s and 16s rRNA), 13 protein-coding genes (PCGs), and 19 transfer RNA (tRNA) genes while tRNA-Leu, tRNA-Cys, and tRNA-Tyr were not characterized, compared with the mitogenome of Thermophis zhaoermii. The typical ATN (ATG, ATT, ATA, or ATC) start codons and T++ stop codons are present in PCGs with the exception of start codon GTG and stop codon AG + for COX1 and ND4. The sequence length of 12s and 16s rRNA were 919 bp and 1468 bp, respectively, which were separated by tRNA-Val. The phylogenetic tree () obtained from bayesian inference (BI) strongly indicated that Pseudoxenodon bambusicola is embedded in the Colubridae family. This resource is the first complete mitochondrial genome of snakes of the subfamily Pseudoxenodontinae and will contribute for further comparative mitogenome studies as well as the evolutionary relationships within Colubridae families.
Figure 1. Phylogenetic tree of the relationships among snakes based on Bayesian inference (BI) method. The number nearby the node is the value of Bayesian posterior probabilities. The species selected and corresponding GenBank accession number are shown as follows: Pseudoleptodeira latifasciata (NC_013981), Sibynophis chinensis (NC_022430), Sibynophis collaris (NC_016424), Thermophis shangrila (NC_035058), Thermophis baileyi (NC_035713), Thermophis zhaoermii (GQ166168), Xenopeltis unicolor (NC_007402), Oligodon ningshaanensis (NC_026083), Dinodon rufozonatum (KF148622), Elaphe poryphyracea (NC_012770).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Akaike H. 1974. A new look at the statistical identification model. IEEE Trans Autom Contr. 19:716–723.
- Bernt M, Donath A, J uhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Paszkiewicz K, Studholme DJ. 2010. De novo assembly of short sequence reads. Brief Bioinformatics. 11:457–472.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Xia X. 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol. 35:1550–1552.
- Zhang B, Huang S. 2013. Relationship of Old World Pseudoxenodon and New World Dipsadinae, with comments on underestimation of species diversity of Chinese Pseudoxenodon. Asian Herpetol Res. 4:155–165.
