Abstract
Rhabdophis himalayanus listed as a vulnerable species has been included in the IUCN Red List. Here, the complete mitochondrial genome sequence of this kind of snake is first reported. The genome is 17,149 bp in length, containing 2 ribosomal RNA (rRNA) genes, 13 protein-coding genes, 22 transfer RNA (tRNA) genes, and 2 D-loop regions. The mitochondrial genome order, nucleotide composition, and codon usage pattern of it are similar to other snakes. The phylogenetic results indicating that R. himalayanus can be firmly placed in the family Colubridae, under the genus Rhabdophis.
Rhabdophis himalayanus (CitationGünther, 1864) was included in the ‘National Protected List of Terrestrial Wild Animals with Good Benefits or Important Economic and Scientific Values’ issued by the State Forestry Administration of China on 1 August 2000, and in the Chinese Red List of Biodiversity - Vertebrates Volume, assessed as Vulnerable (VU), and included in the IUCN in 2013. The morphological characteristics, distribution (Zhao Citation2006), hemipenial morphology (Zhu et al. Citation2013) explored the validity of the R. himalayanus. However little is known about the complete mitochondrial DNA of this species.
A specimen of Rhabdophis himalayanus (SICAU15008, stored in the zoological specimens room, College of Life Science, Sichuan Agricultural University) was collected from Tibet, China and preserved with 95% alcohol. Total DNA was extracted from the muscle using a Genomic DNA Extraction Kit according to the instruction. The Complete mitochondrial genome was amplified using PCR with 22 pairs of primers, overlapping fragments covering the complete mitochondrial genome of R. himalayanus. PCR products were purified and sequenced by the company (TsingKE) by using the same primers. The mitogenome sequence and protein-coding genes were assembled and analyzed by MEGA 5.0 (Tamura et al. Citation2011). Phylogenetic analysis by BI was carried out using MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and ML analysis was performed using PAUP4.0b10 (Swofford Citation2003).
The complete mitochondrial genome of Rhabdophis himalayanus is 17,149 bp in length, It contained 37 typical mitochondrial genes (13 protein-coding genes, 22 tRNAs and 2 rRNAs) and 2 control regions (D-loop1 and D-loop2). The total base composition of the mitochondrial genome is 26.36% T, 26.90% C, 33.98% A, and 12.76% G. Most genes of R. himalayanus were encoded on the H-strand, in addition to the ND6 and nine tRNA genes: tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer (UCN), tRNAGlu, tRNAThr, and tRNAPro, which were encoded on the L-strand. Among the 13 protein-coding genes, the longest gene was ND5 (1761 bp), while the shortest gene was ATP8 (165 bp). The 12S rRNA was located between tRNAPhe and tRNAVal, while 16SrRNA was located between tRNAVal and ND1. D-loop 1 was located between tRNAPro and tRNAphe and 1045 bp in length, while D-loop 2 was located between tRNAIle and tRNALeu (UUR), and 989 bp in length. The base composition of the two control regions was very similar.
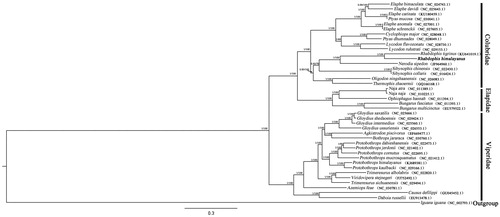
In this study, the phylogenetic relationships are based on the concatenated sequences of 12 heavy-strand protein-coding genes (). The maximum likelihood (ML) and Bayesian inference (BI) constructed have the same topology. Rhabdophis himalayanus and Rhabdophis tigrinus formed a monophyletic group (PP = 1, BS = 100), confirm in the validity of Rhabdophis himalayanus placed under the genus Rhabdophis (Zhao Citation2006; Zhao et al. Citation2016). This is the first time to reveal the complete mitochondrial genome of R. himalayanus. This not only can provide important data for the phylogenetic evolution research and genetic identification of vertebrates, including snakes, but also contribute to the conservation of this endangered wild animal’s genetic resources.
Figure 1. Bayesian tree and maximum likelihood tree estimated using 12 protein-coding genes alignment. The phylogenetic tree shown is based on the Bayesian tree topology, adding the bootstrap and posterior probability values of the nodes (BS/PP). The Genbank accession numbers for the samples are shown in the tree. Rhabdophis himalayanus appears in bold.
Acknowledgements
We thank Dr. Li Ding for his assistance in sample collection.
Disclosure statement
The authors declare that they have no competing interests. The authors alone are responsible for the writing of this paper.
Additional information
Funding
References
- Günther A. 1864. The Reptiles of British India. Ray Society by Robert Hardwicke, London: p. 544.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sunder land (MA): Sinauer Associates.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Zhao EM. 2006. The snakes of China [in Chinese] Vol. I. Hefei, China: Anhui Sience & Technology Publ. House; p. 372.
- Zhao D, Liu H, Zhao WG, Liu P. 2016. The complete mitochondrial DNA sequence and the phylogenetic position of Rhabdophis tigrinus (Reptilia: Squamata). Mitochondrial DNA Part B Resources. 1:216–217.
- Zhu GX, Guo P, Zhao EM. 2013. Comparative studies on hemipenial morphology of eight species of Keelback snakes (Serpentes: Colubridae: Rhabdophis). Sichuan J Zool. 32:380–384.
