Abstract
The complete mitochondrial genome of Otus sunia was 17,835 bp in length and its overall base composition was estimated to be 31.0% for A, 23.3% for T, 32.4% for C, and 13.3% for G. It consisted of the typical structure of 13 PCGs, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 control region. The phylogenetic analysis demonstrated that O. sunia was more closely related to O. scops than O. bakkamoena.
Otus sunia, commonly called oriental scops owl, has been classified as a Class II nationally protected species in China (Holt et al. Citation2018). Therefore, it is crucial to figure out a valid conservation strategy. However, there is very little molecular information about this species, while the mitochondrial genome (mitogenome) sequence is proved to be informative and very useful for systematics and conservation biology studies (Curole and Kocher Citation1999). Particularly, previous studies have demonstrated that molecular markers generated from mitogenome sequence played a vital role in conservation studies and practices for protected animal species (Boore Citation1999; Galtier et al. Citation2009), for example, quick discoveries of illegal trafficking and poaching. In this study, we sequenced and annotated the complete mitochondrial genome of O. sunia first, and examined the phylogenetic position of O. sunia within Strigiformes to provide the basic data for conservation genetics.
Muscle sample of a wild female O. sunia that died of natural causes, was collected from Laojunshan National Nature Reserve, Yibin, Sichuan Province, China (104°00.99’, 28°41.98’). The specimen, at present, is stored in the Museum of Sichuan University. Primers used in this study were designed from the conservative regions on basis of the alignment of complete mitochondrial genomes available within the family Strigidae. Genomic DNA extraction, PCR amplification, sequencing, and annotation were conducted following the methods described by Zhou et al. (Citation2017).
The complete mitochondrial genome of O. sunia has been deposited in the GenBank under the accession number of MF346692. The total length of the complete mitochondrial genome of O. sunia was 17,835 bp, containing 13 protein-coding (PCGs), 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and 1 control region. The overall base composition was 23.3% of T, 32.4% of C, 31.0% of A, and 13.3% of G with an A + T ratio of 54.3%. The nucleotide compositions of the entire O. sunia mtDNA indicate the occurrence of more T and A than G and C (AT skew, 0.14; GC skew, −0.42). Resembling typical avian mtDNA, all the genes of O. sunia mtDNA encoded on the H-strand, apart from one PCG (ND6) and eight tRNAs (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser(UCN), tRNA-Pro, and tRNA-Glu) (Dove et al. Citation2008).
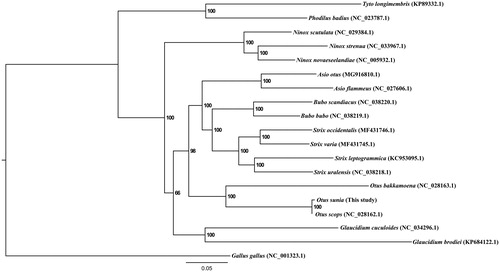
To explore the evolutionary status of O. sunia within the order Strigiformes, a maximum likelihood (ML) tree was constructed based on the concatenated 12 PCGs using RAxML (Stamatakis Citation2014) (). Gallus gallus was used as the outgroup for tree rooting. The phylogenetic tree showed that O. sunia was more closely related to O. scops than O. bakkamoena. In conclusion, our study describes the complete mitogenome of O. sunia, and defines its phylogenetic position, which would facilitate further investigations of molecular evolution and conservation of this species.
Acknowledgements
We are thankful to Guo Cai for collecting the specimen.
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Curole JP, Kocher TD. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. (Amst.). 14:394–398.
- Dove CJ, Rotzel NC, Heacker M, Weigt LA. 2008. Using DNA barcodes to identify bird species involved in birdstrikes. J Wildl Manage. 72:1231–1236.
- Galtier N, Nabholz B, Glémin S, Hurst GD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541–4550.
- Holt DW, Berkley R, Deppe C, Enríquez Rocha P, Petersen JL, Rangel Salazar JL, Segars KP, Wood KL, Kirwan GM, Marks JS. 2018. Oriental Scops-owl (Otus sunia). In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E. editors. Handbook of the birds of the world alive. Barcelona: Lynx Edicions. (retrieved from https://www.hbw.com/node/54960 on 1 October 2018)
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zhou C, Hao Y, Ma J, Zhang W, Chen Y, Chen B, Zhang X, Yue B. 2017. The first complete mitogenome of Picumnus innominatus (Aves, Piciformes, Picidae) and phylogenetic inference within the Picidae. Biochem Syst Ecol. 70:274–282.