Abstract
In the present study, we reported the complete mitochondrial DNA sequence of the hybrid from wild yak (♂) × domestic yak (♀). The mitochondrial genome is 16,322 bp in length, including the typical structure of 22 transfer RNA genes, 13 protein-coding genes, 2 ribosomal RNA genes, and 1 non-coding control region. The overall nucleotide composition is: 33.74% A, 13.18% G, 25.86% C, and 27.21% T. The phylogenetic analysis by neighbor-joining (NJ) method showed that the hybrid of wild yak (♂) × domestic yak (♀) has a closer relationship to the Qinghai Plateau yak.
The wild yak (Bos mutus) is rare, yet, iconic bovine species inhabiting the Tibetan Plateau. Its range include the western edge of Gansu Province, Qinghai Province, the southern rim of the Xinjiang Autonomous Region, and the Tibet Autonomous Region (Schaller and Liu Citation1996). The number of wild yaks declined in the 20th century, as a result of excessive human hunting and habitat destruction. At present, the wild yak is categorized as State Class I (the highest class) protected animal in China, and Vulnerable on the IUCN Red List (Harris and Leslie Citation2008). Recent studies have found that hybridization between wild and domestic yak could heighten the genetic contamination of wild yak populations (Leslie and Schaller Citation2009). The growth performance and stress tolerance of the hybrids between domestic and wild yak were better than those of domestic yak at the same age (Wiener et al. Citation2003). Some pastoralists drive their female domestic yaks into regions where wild yaks lived during the breeding season, in order to allow natural mating with wild yak bulls. In the face of hybridization, it is necessary to evaluate the genetic information of pure wild yak individuals and hybridized individuals. Mitochondrial DNA (mtDNA) is one of the most useful molecular tools in biodiversity conservation and population genetics (Stephen Citation2014). However, there is little information about the complete genome of the wild yak (♂) × domestic yak (♀)
In this study, a specimen of the hybrid individual of wild yak (♂) × domestic yak (♀) was obtained from Tianjun County, Qinghai Province, China (N37°18′1.98″, E99°01′18″) and the blood sample was stored in the Key Laboratory of Yak Breeding Engineering of Gansu Province. Total genomic DNA was extracted from the individual blood samples using the genomic DNA isolation kit (TIANGENE, Beijing, China) according to the manufacturer’s instructions. Six primers were designed to amplify the PCR products for sequencing. The complete mitochondrial DNA sequence of the hybrid from wild yak (♂) × domestic yak (♀) was determined and deposited in GenBank with accession number MF973066.1.
The complete mitochondrial genome of hybrid is 16,322 bp in length with the typical gene order and transcriptional direction of vertebrates. It consists of 37 mitochondrial genes, including 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes (12S and 16S rRNA), and 1 D-loop region. The overall nucleotide composition is 33.74% A, 13.18% G, 25.86% C, and 27.21% T, with a total A + T content of 60.95%.
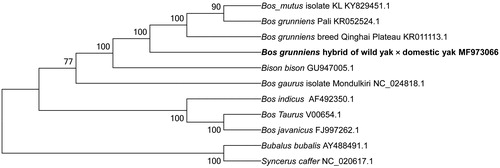
To validate the phylogenetic position of hybrid, we constructed a neighbor-joining (NJ) tree with 1000 bootstrap replicates by MEGA 6.0 (Tamura et al. Citation2013) based on 11 complete mitochondrial genomes of Bovidae. As shown in , the hybrid has the closest relationship to the Qinghai Plateau yak. The complete mitochondrial genome of the hybrid yak will provide fundamental information for effective management and conservation strategy development for the wild yak.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Harris RB, Leslie D. 2008. Bos mutus. In The IUCN Red List of Threatened Species. e.T2892A9492411.
- Leslie DM, Schaller GB. 2009. Bos grunniens and Bos mutus (Artiodactyla: Bovidae). Mamm Spec. 836:1–17.
- Schaller GB, Liu W. 1996. Distribution, status, and conservation of wild yak Bos grunniens. Biol Conserv. 76:1–8.
- Stephen L. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–91.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wiener G, Han J, Long R. 2003. The yak, 2nd edn. Bangkok, Thailand: Regional Office for Asia and the Pacific Food and Agriculture Organization of the United Nations.