Abstract
The complete mitochondrial genome (mitogenome) of Dnopherula yuanmowensis was sequenced and characterized in our study. The mitogenome is a circular molecule of 15,629 bp in length, containing 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and an A + T-rich region, as in other grasshoppers. The AT content of the overall base composition is 74.9%. Phylogenetic reconstruction using both Bayesian Inference (BI) and Maximum Likelihood (ML) validated the taxonomic status of D. yuanmowensis, placing it in the monophyletic Gomphocerinae.
Dnopherula yuanmowensis, belongs to the subfamily Gomphocerinae, within the family Acrididae of the order Orthoptera (Eades and Otte Citation2018). Due to the limited effective molecular data, there is no mitogenomic information about this species. Thus, we determined and characterized the complete mitochondrial DNA sequence of D. yuanmowensis, which is the first one for the genus, contributing to aid further phylogenetic and genetic studies of this species.
All specimens were collected in Hongyuan, China. The collected specimens were stored in 95% ethanol and deposited in the College of Plant Protection, Nanjing Agricultural University, Nanjing, Jiangsu. Whole genomic DNA was extracted from hind femoral muscles of each specimen using a Wizard® Genomic DNA Purification Kit (Promega, Madison, USA) according to the manufacturer’s instructions. Certain pairs of universal primers for grasshopper mitochondrial genomes were used for polymerase chain reaction (PCR) amplification (Simon et al. Citation2006). Then PCR products were sequenced using primer-walking strategy from both strands by Genscript Biotech Corp. (Nanjing, China). Mitochondrial genome was assembled by SeqMan program from DNASTAR (Burland Citation2000), and annotated using MITOS Web Server (Bernt et al. Citation2013).
The complete mitogenome of D. yuanmowensis was 15,629 bp in length (GenBank accession no. KY747520). it contained the entire set of 37 genes: 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, and an A + T-rich region (Boore Citation1999). The overall nucleotide composition was 42.67% of A, 10.67% of G, 32.23% of T, and 14.43% of C, with the AT-rich feature (74.90%). The orientation and gene order of the mitogenomes of this species was identical to that of Drosophila yakuba, except that the tRNALys–tRNAAsp could be recognized in Acridoidea rather than the tRNAAsp–tRNALys in D. yakuba (Rozas et al. Citation2017). All protein-coding sequences had a typical ATN codon. Twelve PCGs used a common stop codon of TAN (10 with TAA and 2 with TAG), whereas the remaining one (ND5) was terminated by the incomplete stop codon T. The 22 typical tRNA genes were interspersed in the genome and range from 63 to 72 bp in length. The 12S and 16S rRNA genes were 843 bp and 1315 bp in length, respectively. The A + T-rich region of the mitogenome was located in the conserved position between the 12S and tRNAIle with the length of 732 bp.
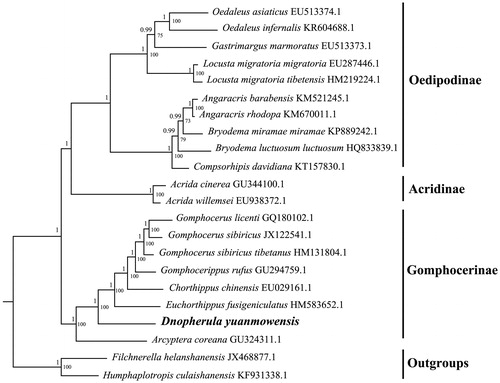
To validate the phylogenetic position of D. yuanmowensis, the ML and BI trees were constructed on CIPRES Portal using 13 mitochondrial PCGs from mitogenomes of 20 species, and two outgroups from Pamphagidae. We used the best-fit partitioning scheme and partition-specific models recommended by PartitionFinder (Lanfear et al. Citation2012). Two phylogenetic analyses yielded the same topology, and nodal supporting values were always higher for BI trees than for ML trees, which was often the case in previous studies of many other taxa (Yuan et al. Citation2016; Chen et al. Citation2018). Our analyses showed that species of three subfamilies were clustered as three monophyletic groups. And D. yuanmowensis was positioned in the subfamily of Gomphocerinae (posterior probabilities = 1, bootstrap value = 100%; ). It is apparent that dense taxonomic sampling of Gomphocerinae is in demand to resolve the phylogenetic position of this group of grasshoppers.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for this paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Chen ZT, Zhao MY, Xu C, Du YZ. 2018. Molecular phylogeny of Systellognatha (Plecoptera: Arctoperlaria) inferred from mitochondrial genome sequences. Int J Boil Macromol. 111:542–547.
- Eades DC, Otte D. 2018. Orthoptera species file 5.0/5.0, Available from http://orthoptera.speciesfile.org.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34:3299–3302.
- Simon C, Buckley T, Frati F, Stewart J, Beckenbach A. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Syst. 37:545–579.
- Yuan ML, Zhang QL, Zhang L, Guo ZL, Liu YJ, Shen YY, Shao R. 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol Phylogenet Evol. 104:99–111.