Abstract
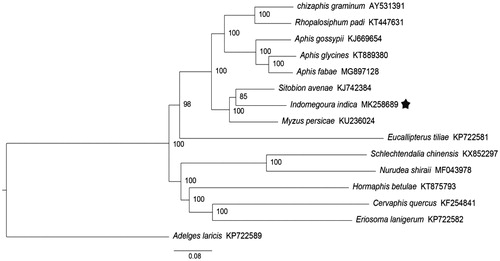
In this study, the complete mitochondrial genome sequence of Indomegoura indica(Hemiptera: Aphididae) was determined. The circular genome was 15220 bp in length and contained 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and two non-coding regions (control regions). All PCGs were initiated with standard ATN codons and terminated with TAA except for cox1 and nad4. All tRNAs were predicted to contain typical cloverleaf secondary structures except trnS1. The Bayesian phylogenetic analysis indicated that Indomegoura indica was more closely related to Sitobion avenae than to other species.
Indomegoura indica (van der Goot) is one kind of aphids which mainly feeds on Hemerocallis spp. plants such as Hemerocallis citrina and Hemerocallis fulva (Zhang and Qiao Citation1998). So far, it is distributed in China (Fujian, Guizhou, Beijing, Taiwan, Jiangsu, and Hebei), Korea, and India (Zhang and Qiao Citation1998; Lee et al. Citation2011; Xu Citation2017). In this study, we sequenced and annotated the complete mitogenome of Indomegoura indica on an Illumina platform (Coil et al. Citation2015). The mitogenome sequence assembly used the grain aphid species Sitobion avenae as the reference genome. After adult specimens were collected from Hemerocallis citrina in Nanzheng County (32°48’N, 106°39’E, altitude 1523 m), Shaanxi, China, they were deposited in the State Key Laboratory of Crop Stress Biology in Arid Areas, College of Plant Protection, Northwest A&F University (NWAFU), Shaanxi, China.
The circular mitochondrial genome of Indomegoura indica was 15,220 bp in length (GenBank accession number MK258689) and contained 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and 2 control regions (non-coding AT-rich regions). The order and orientation of the mitochondrial genes were identical to that of Sitobion avenae. Besides the two AT-rich regions (282 bp and 438 bp, respectively) with A + T contents of 86.25%, there were 15 intergenic spacer regions ranging in size from 1 to 52 bp (114 bp in total) and 6 overlapping regions (34 bp in total) throughout the whole genome.
The nucleotide composition was significantly biased (45.85%, 38.02%, 10.35%, and 5.78%, respectively) with A + T contents of 83.87%. All PCGs were initiated with typical ATN codons (six with ATT, three with ATA, two with ATG, and two with ATC). Whereas 11 PCGs were terminated with TAA, and 2 PCGs were terminated with an incomplete stop codon T (cox1 and nad4). All of 22 tRNAs ranged from 61 to 73 bp and were predicted to contain typical cloverleaf secondary structures except the gene trnS1 (AGN), whose DHU arm was replaced by a simple loop. rrnL was 1265 bp in length with A + T contents of 85.14%, and rrnS was 764 bp in length with A + T contents of 83.64%.
We used the concatenated nucleotide sequences of 13 PCGs from 14 species in the superfamily Aphidoidea and outgroup species Adelges laricis in the superfamily Phylloxeroidea to estimate the phylogenetic analysis. The mitochondrial sequences of these species were downloaded from GenBank of NCBI. Bayesian analyses were performed with MrBayes 3.2.6 (Ronquist and Huelsenbeck Citation2003), and the BI phylogenetic tree showed that Indomegoura indica was more closely related to Sitobion avenae than to other species ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Lee W, Kim H, Lim J, Choi HR, Kim Y, Kim YS, Ji JY, Foottit RG, Lee S. 2011. Barcoding aphids (Hemiptera: Aphididae) of the Korean Peninsula: updating the global data set. Mol Ecol Resour. 11:32–37.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Xu ZH. 2017. How to control Indomegoura indica. Applicable Technologies for Rural Areas. 9:45–46.
- Zhang GX, Qiao GX. 1998. A study on Genus Indomegoura (Hemiptera: Aphididae) and notes on new species. Entomologia Sinica. 5:95–100.