Abstract
Lippia origanoides (Kunth), also known as salva-de-marajó, is an aromatic shrub distributed from central America to northern South America that presents biomedical potential. Here we sequenced a partial genome dataset for salva-de-marajó using 1/11th of a lane on Illumina Hiseq. The software MIRA was used to assemble the complete plastid using different k-mer sizes and further merging well-assembled regions through manual curation. The plastid genome was 154,310 bp in length, divided in one Large Single Copy region containing 85,156 bp; one Small Single Copy region containing 13,320 bp; and two inverted repeat regions presenting different sizes: IRa, containing 25,955 bp; and IRb, containing 29,879 bp. Genome annotation was automatically performed using GeSeq. Artemis was used to perform manual curation. The plastid presents 88 protein-coding genes, 36 tRNAs and 8 rRNAs. It was deposited in GenBank under the accession number MK248831. Twenty-three species of Lamiales containing complete plastids distributed along nine different families were downloaded to confirm the phylogenetic position of this newly described genome, plus 3 outgroup species. Forty-three genes were found to be present in all plastids. These genes were aligned and concatenated to produce a dataset of 13,959 nucleotidic positions. Modeltest defined the best substitution model as the General Time Reversible. As expected, the phylogenomic analysis grouped L. origanoides together with Aloysia citrodora in a monophyletic clade representing the Verbenaceae family. The complete chloroplast genome of L. origanoides will contribute to support further studies on population genetics, phylogeography, evolutionary, and conservation biology of Verbenaceae.
Lippia origanoides, Verbenaceae, is native to some countries from Central and northern South America (Pascual et al. Citation2001). This shrub is also named salva-de-marajó and has widespread use in folk medicine (Oliveira et al. Citation2014), being present in the priority list of plant species from the Brazilian Government (SUS) and recommended by the Brazilian Pharmacopoeia. Its essential oil has antimicrobial (Oliveira et al. Citation2007) and antifungal activity (Betancur et al. Citation2011). It also presents potential as a natural pest controlling agent (Mar et al. Citation2018), a preservative for food, and use in pharmaceutical and cosmetic products (Hernandes et al. Citation2017).
Aerial parts of L. origanoides were collected from a cultivated specimen at the city of Oriximiná (Pará State, Brazil, 16°36’15,1”S and 49°16’0,70”W). A specimen was deposited at UFJF Herbarium under the voucher CESJ 39532. After DNA extraction, a paired-end genome library was produced and sequenced using Illumina Hiseq platform, generating 85,724,792 sequencing reads.
These reads were assembled de novo using MIRA software (Chevreux et al. Citation1999) with different k-mer sizes. The biggest contigs were ordered by comparison against Aloysia citrodora plastid (NC_034695) to produce a guide sequence used as a backbone to new reference-based assemblies with different k-mers. The well-assembled regions were defined by checking the read overlapping on Tablet software (Milne et al. Citation2013) and used to build a second guide sequence. Some remaining gaps were closed using in silico PCR approaches.
The complete chloroplast genome of L. origanoides (MK248831) presents a total length of 154,310 bp. It consists of a Large Single Copy region, a Small Single Copy region and two Inverted Repeat regions. As observed in A. citrodora and possibly representing a feature of Verbenaceae plastids, a longer IR presented 29,879 bp and a shorter one contained 25,955 bp. The shorter IR presents a truncated version of the ycf1 gene.
A total of 554,253 reads were used to assemble the complete chloroplast genome with an average read coverage of 1019×. The overall GC content was 39%. The chloroplast genome of L. origanoides was automatically annotated using GeSeq (Tillich et al. Citation2017) and manually curated using Artemis (Carver et al. Citation2012). It contained 132 annotated features, being 88 protein-coding genes, 36 tRNAs, and 8 rRNAs.
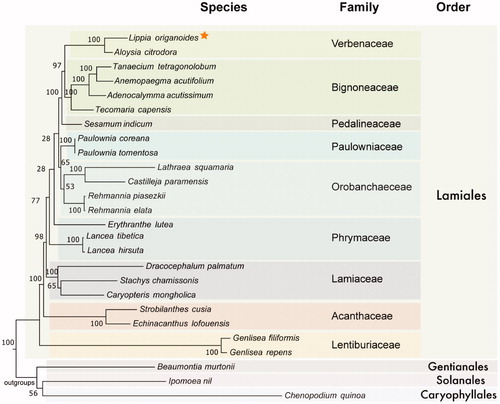
In order to confirm the phylogenetic position of this newly described genome, we downloaded the data from 23 complete plastids from the order Lamiales and three outgroups. We chose plastids from nine different families and used 43 genes contained in all samples to produce a dataset of 13,959 nucleotides. The genes were aligned and then concatenated. A phylogenomic analysis () was performed using maximum likelihood under MEGA7 (Kumar et al. Citation2016). As expected, all genera were recovered as monophyletic as well as all families, with the exception of Phrymaceae that showed low statistical support. Lippia origanoides was grouped in the monophyletic family Verbenaceae together with Aloysia citrodora. The complete chloroplast genome of the Salva-de-marajó is an important resource that will contribute to further studies on population genetics, phylogeography, evolutionary, and conservation biology of Verbenaceae.
Figure 1. Evolutionary history of Lamiales inferred by Maximum Likelihood based on the General Time Reversible model using a total of 43 genes (13,959 positions) shared between 23 species of Lamiales and three outgroups. All positions containing gaps and missing data were eliminated. Bootstrap values (1000 replicates) are shown in the corresponding nodes. Orange star highlights the plastid produced here. Colored boxes were used to cluster members from different families and orders. Plastid accession numbers are: MK248831, NC_034695, NC_027955, NC_037226, NC_037455, NC_037462, NC_016433, NC_031435, NC_031436, NC_027838, NC_031805, NC_034311, NC_034312, NC_030212, NC_037693, NC_037506, NC_031874, NC_029822, NC_035729, NC_037485, NC_035876, NC_037079, NC_037081, NC_037934, NC_031159, NC_034949.
Acknowledgements
We are thankful to Maria Glace Nery Cardoso Nunes for supplying the plant specimen, Dr. Fátima Regina Gonçalves Salimena for sample identification.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Betancur L, Zapata B, Baena A, Bueno J, Nova C, Stashenko E, Mesa A. 2011. Antifungal, cytotoxic and chemical analyses of essential oils of Lippia origanoides H.B.K grown in Colombia. Revista Salud UIS. 43:141–148.
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis:an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 28:464–469.
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Computer science and biology: proceedings of the German conference on bioinformatics (GCB) 99:45–56.
- Hernandes C, Pina ES, Taleb-Contini SH, Bertoni BW, Cestari IM, Espanha LG, Varanda EA, Camilo KFB, Martinez EZ, França SC, et al. 2017. Lippia origanoides essential oil: An efficient and safe alternative to preserve food, cosmetic and pharmaceutical products. J Appl Microbiol. 122:900–910.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Mar J, Silva L, Azevedo S, França L, Goes A, Dos Santos A, Araujo Bezerra J, De Cássia R, Nunomura S, Machado M, et al. 2018. Lippia origanoides essential oil: an efficient alternative to control Aedes aegypti, Tetranychus urticae and Cerataphis lataniae. Ind Crops Prod. 111:292–297.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using tablet for visual exploration of second-generation sequencing data. Briefings in Bioinformat. 14:193–202.
- Oliveira D, Leitão G, Bizzo RH, Lopes D, Alviano DS, Alviano CS, Leitão SG. 2007. Chemical and antimicrobial analyses of essential oil of Lippia origanoides H.B.K. Food Chem. 101:236–240.
- Oliveira D, Leitão G, Fernandes P, Leitão S. 2014. Ethnopharmacological studies of Lippia origanoides. Rev Bras Farmacogn. 24:206–214.
- Pascual ME, Slowing K, Carretero E, Mata DS, Villar A. 2001. Lippia: traditional uses, chemistry and pharmacology: a review. J. Ethnopharmacol. 76:201–214.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
