Abstract
Male-only rearing has long been desirable in sericulture because males have superior economic characteristics. To this end, a balanced sex-linked lethal (BSL) system has been developed. Here, we report the first complete mitochondrial genome of the BSL silkworm (Bombyx mori). Phylogenetic analysis revealed that the BSL silkworm is more closely related to Japanese B. mori than Chinese B. mori. The new mitochondrial genome provides valuable information to further explore genetic diversity and origin of this species.
As the foundation of sericulture, the mulberry silkworm, Bombyx mori, has been used for silk production for thousands of years (Goldsmith et al. Citation2005). Male-only rearing has long been sought in sericulture because males have superior economic characteristics, such as higher silk yield, higher silk quality, and lower food consumption (Xia and Tang Citation1980). To this end, Strunnikov (Citation1975) first established a balanced sex-linked lethal (BSL) system by classic genetics. The BSL system was then introduced to our institute in the last century and applied in Chinese sericulture. For a better understanding of its genetic diversity and phylogenetic position, we analyzed the complete mitochondrial genome of the BSL silkworm, called as B. mori Ping1 in China.
B. mori Ping1 was collected from Hangzhou City, Zhejiang Province in China (30°18′ N, 120°11′ E). The specimens were stored in the Silkworm Germplasm Resources Repository at Sericulture Research Institute, Zhejiang Academy of Agricultural Sciences. Total genomic DNA was extracted from a single pupa using a DNeasy Tissue Kit (QIAGEN, Hilden, Germany). Whole mitochondrial genome was determined using Sanger sequencing. The DNA sequence was analyzed using MITOS (Bernt et al. Citation2013) and tRNAscan-SE server (Schattner et al. Citation2005). The complete mitogenome is 15,675 bp in length (GenBank accession no. MK246423) and contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 non-coding control region. The overall base composition was 43.10% A, 38.31% T, 11.30% C, and 7.29% G, with a high A + T content of 81.41%.
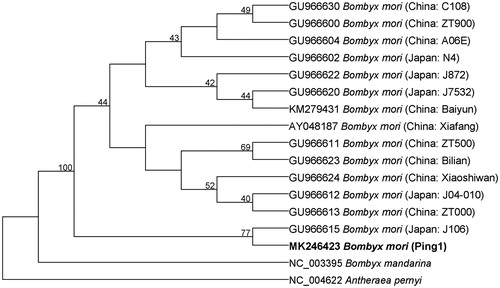
To validate the phylogenetic position of B. mori Ping1, we used MEGA 7.0 (Kumar et al. Citation2016) to construct a maximum-likelihood tree based on the complete mitochondrial DNA sequence. Bombyx mandarina and Antheraea pernyi were used as outgroups. As shown in the phylogenetic tree (), B. mori Ping1 is more closely related to Japanese B. mori J106 than Chinese B. mori, inconsistent with the result of our observation on their cocoon shapes. B. mori Ping1 had been thought to be Chinese silkworm strain because they both spin a globular or spherical cocoon, in contrast with a peanut-shaped cocoon by Japanese silkworm strain (Kiyosawa et al. Citation1999). Our finding suggests that the maternal ancestor of B. mori Ping1 comes from Japanese silkworm strain, while its paternal ancestor originates from Chinese silkworm strain. More work will be needed to further confirm this finding.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Goldsmith MR, Shimada T, Abe H. 2005. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 50:71–100.
- Kiyosawa M, Ito E, Shirai K, Kanekatsu R, Miura M, Kiguchi K. 1999. Cocoon spinning behavior in the silkworm, Bombyx mori: comparison of three strains constructing different cocoons in shape. Zool Sci. 16:215–223.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan, and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686.
- Strunnikov VA. 1975. Sex control in silkworms. Nature. 255:111–113.
- Xia JG, Tang WY. 1980. The relationship between sex and economic characteristics in silkworm. Acta Sericologica Sinica. 6:167–172.