Abstract
Complete chloroplast genomes were characterized for two economically important tree species, Cinnamomum camphora and Cinnamomum parthenoxylon. The chloroplast genomes are 152,721 bp (C. camphora) and 152,760 bp (C. parthenoxylon) long, respectively, and both comprise a pair of inverted repeat (IR) regions, separated by a large single-copy (LSC) region and a small single-copy (SSC) region. The same panel of 112 gene species were annotated for both species, including 78 protein-coding (PCG), 30 tRNA, and 4 rRNA gene species. Phylogenetic analysis suggests that the species delimitation and interrelationship of C. camphora and C. parthenoxylon may need further investigation.
Cinnamomum is a genus of evergreen aromatic trees and shrubs within the family Lauraceae, and comprises about 350 species occurring in tropical and subtropical Asia, Australia, and Pacific islands (Li et al. Citation2008; Huang et al. Citation2016). Here, we present the complete chloroplast (cp) genomes for two economically important trees within this genus: C. camphora and C. parthenoxylon (hereinafter abbreviated as CC and CP, respectively).
Leaf samples of CC and CP were collected from Xi’an Botanical Garden (Xi’an, China) and Lushan Botanical Garden (Jiujiang, China), respectively. Voucher specimens are held in the College of Life Sciences, Shaanxi Normal University. Total genomic DNAs were extracted using the DNeasy Plant Mini Kit (Qiagen, CA, USA). Following the preparation of shotgun libraries, high-throughput sequencing was conducted with the Illumina HiSeq XTM Ten Sequencing System (Illumina, CA, USA). After trimming with Trimmomatic v0.36 (Bolger et al. Citation2014), 12.6 M and 4.6 M of 150-bp paired raw reads were employed for assembling the cp genomes using MITObim v1.9 (https://github.com/chrishah/MITObim) (Hahn et al. Citation2013), with that of Cinnamomum micranthum (KR014245) (Wu et al. Citation2016) as the reference. Genome annotation was done with the CpGAVAS web server (Liu et al. Citation2012), and was delicately adjusted by comparing with those annotated cp genomes of their congeners (Wu et al. Citation2016; Chen et al. Citation2017; Wu C-S et al. Citation2017).
The circular genomes were determined to be 152,721 bp (CC; MH050970) and 152,760 bp (CP; MH050971) long, respectively, and both comprise a pair of inverted repeat (IR) regions (CC: 20,114 bp; CP: 20,132 bp), separated by a large single-copy (LSC) region (CC: 93,627 bp; CP: 93,636 bp) and a small single-copy (SSC) region (CC: 18,854 bp; CP: 18,860 bp). These two genomes possess almost identical biased nucleotide compositions (30.0% A, 19.6% C, 19.5% G & 30.9% T) with an overall A + T content of 60.9%. The same panel of 112 gene species were annotated for both species, including 78 protein-coding (PCG), 30 tRNA, and 4 rRNA gene species.
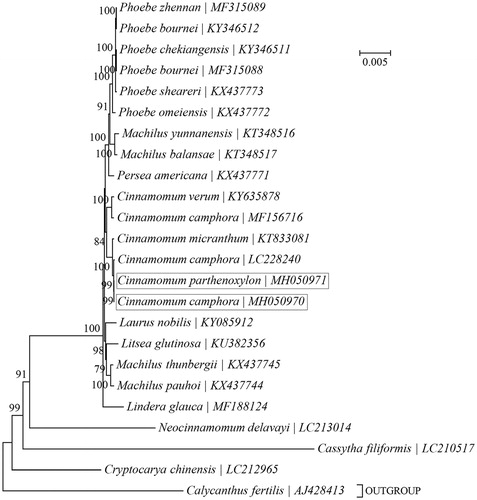
A phylogeny of the family Lauraceae was reconstructed based on the maximum-likelihood (ML) analysis of cp PCGs for a panel of 23 taxa using PhyML-aLRT v2.4.5 (http://www.atgc-montpellier.fr/phyml/alrt/) ) (Guindon and Gascuel Citation2003; Anisimova and Gascuel Citation2006) as in TOPALi v2.5 (http://www.topali.org/) (Milne et al. Citation2009) (). As expected, the five Cinnamomum taxa were more closely related to one another than to the others. However, surprisingly, the cp genome of C. camphora obtained in this study (MH050970) was found to be more closely related to that of C. parthenoxylon than to those two previously reported cp genomes of C. camphora (LC228240; MF156716) (Chen et al. Citation2017; Wu, Wang et al. Citation2017). It should be noted that the two more closely related cp genomes of C. camphora (MH050970; LC228240) possess the identical genomic organization, while the small single-copy (SSC) region of the relatively distant one (MF156716) displays an opposite direction. Our study suggests that the species delimitation and interrelationship of C. camphora and C. parthenoxylon may need further investigation.
Figure 1. Phylogenetic relationships among 23 species within the family Lauraceae based on the maximum-likelihood (ML) analysis of chloroplast PCGs. Calycanthus fertilis (AJ428413) was included as outgroup taxa. ‘GTR + G’ was employed as the best-fit nucleotide substition model. The bootstrap values next to the branches are based on 200 resamplings.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 55:539–552.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Chen C, Zheng Y, Liu S, Zhong Y, Wu Y, Li J, Xu L-A, Xu M. 2017. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PeerJ. 5:e3820.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Huang J-F, Li L, van der Werff H, Li H-W, Rohwer JG, Crayn DM, Meng H-H, van der Merwe M, Conran JG, Li J. 2016. Origins and evolution of cinnamon and camphor: a phylogenetic and historical biogeographical analysis of the Cinnamomum group (Lauraceae). Mol Phylogenet Evol. 96:33–44.
- Li X, Li J, van der Werff H. 2008. Cinnamomum Schaeffer. Flora of China. 7:166–187.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25:126–127.
- Wu C-C, Chu F-H, Ho C-K, Sung C-H, Chang S-H. 2017. Comparative analysis of the complete chloroplast genomic sequence and chemical components of Cinnamomum micranthum and Cinnamomum kanehirae. Holzforschung. 71:189–197.
- Wu C-C, Ho C-K, Chang S-H. 2016. The complete chloroplast genome of Cinnamomum kanehirae Hayata (Lauraceae). Mitochondr DNA. 27:2681–2682.
- Wu C-S, Wang T-J, Wu C-W, Wang Y-N, Chaw S-M. 2017. Plastome evolution in the sole hemiparasitic genus laurel dodder (Cassytha) and insights into the plastid phylogenomics of Lauraceae. Genome Biol Evol. 9:2604–2614.
