Abstract
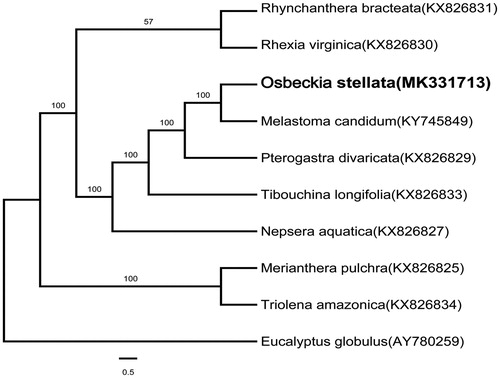
Osbeckia stellata (Melastomataceae) is a shrub distributed in the South. Although many species of Osbeckia have important medicinal value, the phylogenetic position of Osbeckia within the family Melastomataceae remains unclear. Osbeckia stellata is one of the most widespread species in this genus. Here, we reported and characterized the complete chloroplast genome sequence of O. stellata assembled from Illumina sequencing reads. The chloroplast genome size is 156,372 bp, containing a large single copy (LSC) region of 85,741 bp, a short single copy (SSC) region of 17,115 bp, and a pair of inverted repeats (IRs) of 26,758 bp. We predicted a total of 136 genes consisting of 86 protein-coding genes, 48 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that O. stellata was sister to Melastoma candidum.
Osbeckia L. is a genus of herbs, shrublets, or shrubs in the family Melastomataceae, consisting of about 50 species. Osbeckia is widely distributed in tropical West Africa and tropical and subtropical Asia (Chen and Renner Citation2007). Many species in this genus have medical values (Nicholl et al. Citation2001; Syiem and Khup Citation2006). To date, phylogenetic position of Osbeckia in the family Melastomataceae remains unclear. Osbeckia stellata Buchanan-Hamilton ex Kew Gawler is one of the most widely distributed species in this genus, ranging from southern and southwestern China to southeastern Himalayan region (Wu Citation2006; Das and Anjeza Citation2013). Although wildly growing in the field, it is a kind of endophytic fungal medicinal plant that has great potential for clinical use (Bhagobaty and Joshi Citation2012). It was documented that the Apatani tribe, which was located in northeastern India traditionally made use of the leaves of O. stellata to treat toothache (Kala Citation2005) while in Nepal, it was used to heal the wound (Balami Citation2004). Here, we report the complete chloroplast genome sequence of O. stellata to resolve the phylogenetic position of Osbeckia within the Melastomataceae family. The sequence and gene annotation are available in GenBank under accession number MK331713.
One individual of O. stellata was collected in Dhunche, Nepal, and the voucher specimen (Q. Fan17035) was deposited in the Herbarium of Sun Yat-sen University (SYS). DNA was extracted from the dried leaves and a shotgun DNA library was constructed and then sequenced on an Illumina Hiseq X Ten platform (Guangzhou, China). A total of 11.52 Gb paired-end reads were generated and then put into the NOVOPlasty (Dierckxsens et al. Citation2017) to assemble the chloroplast genome with rbcL gene sequence of the same species (GenBank accession number U26330) as a seed. The annotation was performed using the DOGM online tool (Wyman et al. Citation2004) and further manual adjustment was conducted when necessary. For phylogenetic analysis, we used the complete chloroplast genome sequences of O. stellata and eight other species in the family Melastomataceae. Eucalyptus globules from the family Myrtaceae was used as an outgroup. The sequences of 10 species in total were aligned using MAFFT (Katoh and Standley Citation2013) and a maximum likelihood tree was constructed with RAxML implemented in Geneious ver.10.1 (Kearse et al. Citation2012).
The complete chloroplast genome of O. stellata is 156,372 bp in length. It contains two inverted repeats (IRs) of 26,758 bp which seperates the regions of a large single copy (LSC) of 85,741 bp and a small single copy (SSC) of 17,115 bp. The overall GC content is 36.84%. The gene composition of this chloroplast genome we predicted was a total of 136 genes, consisting of 86 protein-coding genes, 48 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that O. stellata was sister to Melastoma candidum with 100% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Balami NP. 2004. Ethnomedicinal uses of plants among the newar community of pharping village of Kathmandu distinct, Nepal. Tribhuvan Univ J. 24:13–19.
- Bhagobaty RK, Joshi SR. 2012. Enzymatic activity of fungi endophytic on five medicinal plant species of the pristine sacred forests of Meghalaya, India. Biotechnol Bioprocess Eng. 17:33–40.
- Chen J, Renner SS. 2007. Melastomataceae. In: Wu ZY, Raven PH, editors. Flora of China. Beijing: Science Press; p. 360–399.
- Das S, Anjeza C. 2013. Antimicrobial and antioxidant activities of Osbeckia stellata Buch.-Dam. ex D.Don (Melastomataceae) prevalent of Darjeeling hills. Int J Pharm Pharm Sci. 5:551–554.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kala CP. 2005. Ethnomedicinal botany of the Apatani in the Eastern Himalayan region of India. J Ethnobiol Ethnomed. 1:11. http://www.ethnobiomed.com/content/1/1/11
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Nicholl DS, Daniels HM, Ira Thabrew M, Grayer RJ, Simmonds MS, Hughes RD. 2001. In vitro studies on the immunomodulatory effects of extracts of Osbeckia aspera. J Ethnopharmacol. 78:39–44.
- Syiem D, Khup PZ. 2006. Study of the traditionally used medicinal plant Osbeckia chinensis for hypoglycemic and anti-hyperglycemic effects in mice. Pharm. Biol. 44:613–618.
- Wu ZY. 2006. Flora of Yunnan (volume1). Beijing: Science Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3325.