Abstract
Forsyth’s toad-headed agama, Phrynocephalus forsythii, is endemic to the Taklamakan Desert. A nearly complete mitochondrial genome of one individual for this species was determined by next-generation sequencing. The mitogenome is 16,060 bp in length, comprising 2 ribosomal RNA genes, 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and a control region (D-loop). The gene arrangement and composition of P. forsythii is identical to the mitogenome of P. theobaldi in that tRNA-Pro was translocated immediately downstream of tRNA-Phe. The D-loop comprised two parts, one (783 bp) existing between tRNA-Thr and tRNA-Phe and another (with 200 bp already sequenced) inserting between tRNA-Pro and 12S rRNA. This mitogenome sequence may provide more data for unveiling the phylogenetic origin and adaptive evolution related to Phrynocephalus viviparity.
The traditional protocols to determine the mitochondrial genomes relied on performing either standard/long PCR or cloning, followed by a series of Sanger sequencing (Kumazawa and Endo Citation2004; Gissi et al. Citation2008). Recent improvements in next-generation sequencing (NGS) technologies have facilitated the obtainment of mitochondrial genomes (Hahn et al. Citation2013; Smith Citation2016).
The Forsyth’s toad-headed agama, Phrynocephalus forsythii, is a viviparous species endemic to the Taklimakan Desert, whereas other viviparous congeneric species occur on the Tibetan Plateau (Guo and Wang Citation2007). The phylogenetic placement of this species is still controversial (Jin and Brown Citation2013; Macey et al. Citation2018; Solovyeva et al. Citation2018). In the present study, we assembled and characterized a mitochondrial genome of P. forsythii by using NGS reads through Roche 454 sequencing platform.
The lizard sample was collected from Ruoqiang County, Xinjiang Uygur Autonomous Region, China. The voucher specimen (No. WGXG08351) was deposited in the herpetological collection at Chengdu Institute of Biology, Chinese Academy of Sciences. Genomic DNA was shipped to Majorbio (Shanghai, China) for library construction and sequencing on Roche 454 FLX platform (Roche, Basel, Switzerland). De novo assembly of clean reads was performed using 454’s Newbler v.2.5 (Branford, Connecticut, USA). After assembly, published sequence of P. forsythii (GenBank accession no. KP126516) was used as query for the reference mitogenome and was BLASTed against assembly using BLASTn (Bethesda, Maryland, USA) (BLAST + v2.2.30) to search for contigs with mitochondrial protein-coding and RNA genes. Genes were annotated with the MITOS (Bernt et al. Citation2013) web server.
The nearly complete mtDNA sequence consists of 16,060 bp and contains 2 ribosomal RNA genes, 13 protein-coding genes (PCGs), and 22 transfer RNA genes (tRNAs). The gene order is identical with some reported cases of congeneric species (Chen et al. Citation2016; Liao and Jin Citation2016; Jin and Brown Citation2018), with the tRNA-Pro gene being translocated immediately downstream of tRNA-Phe gene. The control region consisted of two parts, with part one existing between the tRNA-Thr and tRNA-Phe (783 bp), while another inserting between the tRNA-Pro and 12S rRNA (with 200 bp already sequenced). Most genes are encoded on heavy strand (H-strand) except for ND6 and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser[UGA], Glu, and Pro). Most of the PCGs were initiated with the typical ATG codon, except for ND1 with ATA, ND2 and ND3 with ATT. Meanwhile, most PCGs were terminated with the typical TAA/AGA/AGG codons, except for COII, ATP6, COIII, ND3, and Cyt b with the incomplete termination codon T.
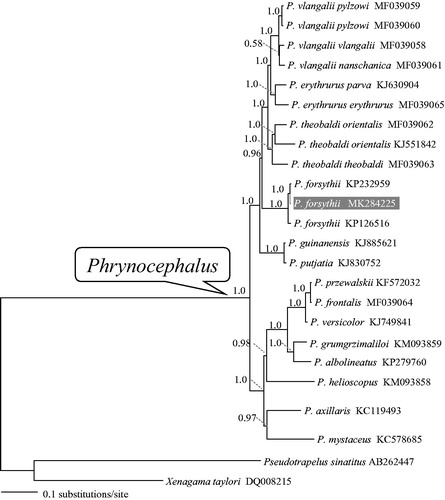
Representative taxa from Phrynocephalus with mitogenome sequences available in GenBank plus two Agaminae taxa as outgroup were used to reconstruct the Bayesian phylogenetic tree for evaluating mitochondrial sequence authenticity of P. forsythii and its phylogenetic placement. The phylogenetic analysis revealed that monophyly of both genus Phrynocephalus and the viviparous group (). Three P. forsythii individuals formed a monophyletic group, sister to the remaining viviparous species exclusive of P. putjatia and P. guinanesis. The mitogenome sequence of P. forsythii will provide more data for resolving evolutionary problems related to viviparity and adaptation of Phrynocephalus.
Figure 1. A majority-rule consensus tree inferred from Bayesian inference using MrBayes v.3.2.1 (Ronquist and Huelsenbeck Citation2003) with GTR + г substitution model, based on the PCGs of 22 individuals of toad-headed lizards and two outgroups. DNA sequences were aligned in MEGA v.6.06 (Tamura et al. Citation2013). The PCGs were translated to amino acids sequences and were manually concatenated all sequences into a single nucleotide dataset (total 11,329 bp). Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution. The novel sequencing sample is highlighted. GenBank accession numbers are given with species/subspecies names.
Nucleotide sequence accession number
The nearly complete mitochondrial genome sequence of P. forsythii has been assigned GenBank accession number MK284225.
Acknowledgements
The authors wish to thank Dr. Yun Xia for help with analysis of mitogenome assembly.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chen D, Zhou T, Guo X. 2016. The complete mitochondrial genome of Phrynocephalus forsythii (Reptilia, Squamata, Agamidae), a toad-headed agama endemic to the Taklamakan Desert. Mitochondrial DNA A. 27:4046–4048.
- Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 101:301–320.
- Guo X, Wang Y. 2007. Partitioned Bayesian analyses, dispersal-vicariance analysis, and the biogeography of Chinese toad-headed lizards (Agamidae: Phrynocephalus): a re-evaluation. Mol Phylogenet Evol. 45:643–462.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Jin Y, Brown RP. 2013. Species history and divergence times of viviparous and oviparous Chinese toad-headed sand lizards (Phrynocephalus) on the Qinghai-Tibetan Plateau. Mol Phylogenet Evol. 68:259–268.
- Jin Y, Brown RP. 2018. Partition number, rate priors and unreliable divergence times in Bayesian phylogenetic dating. Cladistics. 34:568–573.
- Kumazawa Y, Endo H. 2004. Mitochondrial genome of the Komodo dragon: efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 11:115–125.
- Liao P, Jin Y. 2016. The complete mitochondrial genome of the toadheaded lizard subspecies, Phrynocephalus theobaldi orientalis (Reptilia, Squamata, Agamidae). Mitochondrial DNA A. 27:559–560.
- Macey JR, Schulte JA, Ananjeva NB, Dyke ET, Wang Y, Orlov N, Shafiei S, Robinson MD, Dujsebayeva T, Freund GS, et al. 2018. A molecular phylogenetic hypothesis for the Asian agamid lizard genus Phrynocephalus reveals discrete biogeographic clades implicated by plate tectonics. Zootaxa. 4467:1–81.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Smith DR. 2016. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics. 15:47–54.
- Solovyeva EN, Lebedev VS, Dunayev EA, Nazarov RA, Bannikova AA, Che J, Murphy RW, Poyarkov NA. 2018. Cenozoic aridization in Central Eurasia shaped diversification of toad-headed agamas (Phrynocephalus; Agamidae, Reptilia). Peer J. 6:e4543.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
