Abstract
All the genes of Paradoxornis gularis and Niltava davidi complete mitochondrial genomes encoded on the H-strand, apart from one PCG (ND6) and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAPro, and tRNAGlu). Strikingly large tandem repeats were found in the control region (CR) of P. gularis mitogenome. The phylogenetic analysis demonstrated that P. gularis possessed a basal phylogenetic position within the genus Paradoxornis, and N. davidi was more closely related to Cyanoptila cyanomelana than other Muscicapidae species.
Although the whole-genome analyses have achieved rapid advancement and applied in an increasing number of organisms, the popularity of the mitochondrial genome should not be ignored because it is a versatile source of genetic variation for evolutionary study (Wu et al. Citation2015; Zhou et al. Citation2017). There are many evolutionary advantages of mitochondrial genomes, such as freedom from genetic recombination and positive selection, maternal inheritance, and relatively higher mutation rate (Rubtsov and Opaev Citation2012). In recent years, an increasing number of studies have been conducted to expound the molecular phylogenetics in the Passeriformes (Wu et al. Citation2015). To better understand the phylogeny and evolution of the Passeriformes, we determined the complete mitogenomes of P. gularis and N. davidi.
Muscle samples of wild P. gularis and N. davidi that died of natural causes were collected from Laojunshan National Nature Reserve, Yibin, Sichuan Province, China (104°00.99’, 28°41.98’). The specimens are stored in the museum of Sichuan University. Primers used in this study were designed from the conservative regions on basis of the alignment of complete mitochondrial genomes accessible within the Passeriformes. Genomic DNA extraction, PCR amplification, sequencing, and annotation were conducted following the methods described by Zhou et al. (Citation2017).
We have submitted the complete mitogenomes of P. gularis and N. davidi to GenBank under the accession number of KX397391 and KY024217, respectively. The total length of the complete mitochondrial genome of P. gularis and N. davidi were 17,109 bp and 16,770 bp, both containing 13 protein-coding (PCGs), 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes. The overall base composition of P.gularis was 24.0% of T, 31.6% of C, 29.8% of A, and 14.6% of G with an A + T ratio of 53.8%, while 23.7% of T, 31.4% of C, 30.5% of A, and 14.4% of G with an A + T ratio of 54.2% of N. davidi mitogenome. Meanwhile, the AT skew was calculated as 0.107 and 0.125 for the complete mitochondrial genomes of P. gularis and N. davidi. Resembling typical avian mtDNA, all the genes of P. gularis and N. davidi mtDNA encoded on the H-strand, apart from one PCG (ND6) and eight tRNAs(tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAPro, and tRNAGlu) (Dove et al. Citation2008). Particularly important, relatively large tandem repeats (repeat unit: TGTTTTTTCATTTGCTTGACAAAAAAACTAACCATTTTTTCCGACATACCCACAAATTTTTGTTTCGTTCCATTTTTTTTG) in the control region (CR) have been found in P. gularis mitogenome, which was not detected in CRs of other parrotbill mitogenomes.
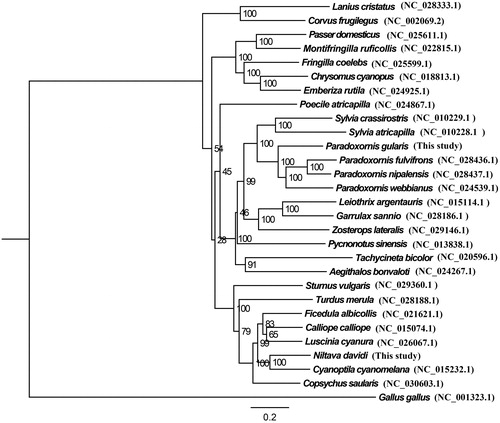
To explore the evolutionary status of P. gularis and N. davidi within the Passeriformes, a maximum likelihood (ML) tree was constructed based on the concatenated 12 PCGs using RAxML (Stamatakis Citation2014) (). Gallus gallus was used as the outgroup for tree rooting. The phylogenetic tree showed that P. gularis possessed a basal phylogenetic position within the genus Paradoxornis, and N. davidi was more closely related to Cyanoptila cyanomelana than other Muscicapidae species. In conclusion, our study described the complete mitogenomes of P. gularis and N. davidi, and defined their phylogenetic position, which would facilitate further investigations of molecular evolution and conservation of the species.
Acknowledgements
We are thankful to Guo Cai for collecting the specimen.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dove CJ, Rotzel NC, Heacker M, Weigt LA. 2008. Using DNA barcodes to identify bird species involved in birdstrikes. J Wildlife Manage. 72:1231–1236.
- Rubtsov AS, Opaev AS. 2012. Phylogeny reconstruction of the yellowhammer (Emberiza citrinella) and pine bunting (Emberiza leucocephala) based on song and morphological characters. Biol Bull Russ Acad Sci. 39:715–728.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wu L, Sun Y, Li J, Li Y, Wu Y, Li D. 2015. A phylogeny of the Passerida (Aves: Passeriformes) based on mitochondrial 12S ribosomal RNA gene. Avian Res. 6:1.
- Zhou C, Hao Y, Ma J, Zhang W, Chen Y, Chen B, Zhang X, Yue B. 2017. The first complete mitogenome of Picumnus innominatus (Aves, Piciformes, Picidae) and phylogenetic inference within the Picidae. Biochem Syst Ecol. 70:274–282.