Abstract
The Formosania is only distributed in a narrow area in Southeast China, and differentiation of the species of this genus may be helpful in understanding the formation process of the water system on both sides of the Taiwan Strait. The complete mitochondrial genome of Formosania chenyiyui is sequenced in this paper. The results show that the full length of the complete mitochondrial genome is 16,572 bp, comprising 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 control region. Overall base composition is 30.35% A, 26.68% T, 15.86% G, and 27.11% C. The result of phylogenetic analysis suggests that F. chenyiyui is close to that of F. lacustre and Vanmanenia stenosoma, and accords with the evolutionary viewpoint of Formosaniini group based on morphological characteristics.
The Gastromyzontidae are a family of loaches native to China and Southeast Asia, and their pectoral and pelvic fins modifed into sucker organs for clinging to objects in fast-flowing streams (Kottelat Citation2012; Nelson et al. Citation2016). The family includes about 125 species in 18 genera. Formosania (Novak et al. Citation2006) is a genus of Gastromyzontidae, which is only distributed in a narrow area on both sides of the Taiwan Straits in Southeast China. The Formosaniini-group (Formerly known as Crossostomini-group) is a special branch in the evolution of the Gastromyzontidae, showing an adaptation to the streams and rivers with a fast current (Fang Citation1935; Chen Citation1980; Tang & Chen Citation2000). At present, nine valid species have been found, and the complete mitochondrial genome of F. lacustre was determined for only one species. Formosania. chenyiyui is distributed only in two rivers, including Hanjiang and Jiulongjiang in Southeast China (Zheng Citation1991). In this study, the complete mitochondrial genome of F. chenyiyui has been determined. Fish specimens were collected from the Tingjiang River (25°57′21.53″ N, 116°19′44.47″ E), Fujian, China. The samples are stored in the Shanghai Ocean University, with an accession number SLFJCT160503009.
The complete mitogenome of F. chenyiyui (GenBank ID: MK135435) is 16,572 bp in length, with overall base composition of 29.70% A, 25.16% T, 16.53% G, and 28.60% C. The circular genome has 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 control region (CR or D-loop). Apart from the ND6 gene and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) encoded on the L-strand, most genes are on the H-strand. There are five overlapping regions with 2–10 bp in length. The genome has 14 intergenic sequences varying from 1 to 31 bp in length. The largest intergenic sequence is located between tRNAAsn and tRNACys. In 13 protein-coding genes, the shortest one is ATP8 gene (168bp) and the longest one is ND5 gene (1830bp). Protein-coding genes use two initiation codons (ATG and GTG). There are four termination codons (TAA, TAG, TA– and T––). The 22 tRNA genes have lengths ranging from 66bp (tRNA Cys) to 76bp (tRNA Lys). The 12S and 16S rRNA genes are 954 and 1680 bp, respectively. D-loop is 916 bp in length.
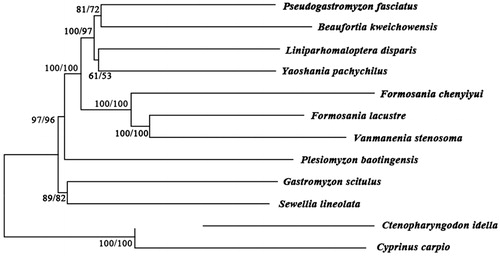
To investigate the phylogenetic relationship about family Gastromyzontidae, we downloaded the mitochondrial genome sequences of nine currently available species of Gastromyzontidae, including Beaufortia kweichowensis (KX060617), Pseudogastromyzon fasciatus (KX101229), Liniparhomaloptera disparis (AP013301), Plesiomyzon baotingensis (KF732713), F. lacustre (AP010774), Vanmanenia stenosoma (NC035628), Yaoshania pachychilus(KT031050), Sewellia lineolate (AP011292), and Gastromyzon scitulus (NC031578), together with Ctenopharyngodon idella (EU391390) and Cyprinus carpio (KF856965) as outgroup species. The phylogenetic trees were constructed using MEGA6 (Tamura et al. Citation2013) for neighbour-joining and maximum-likelihood. Two methods give the same tree topology, and the result indicates that F. chenyiyui is close to that of F. lacustre and V. stenosoma (). The Formosaniini-group contains Formosania and Vanmanenia, which gather into one branch, and then form sister groups with branches of P. fasciatus, B. kweichowensis, Y. pachychilus and L. disparis. This result supports Chen's view about the evolutionary relationship of Formosaniini-group from molecular characteristics (Chen Citation1980).
Figure 1. Phylogenetic tree of family Gastromyzontidae with Ctenopharyngodon idella and Cyprinus carpio as an outgroup. The topology of phylogenetic tree was inferred from neighbour-joining and maximum-likelihood methods. Tree topology was evaluated by 1000 boot-strap replicates. Bootstrap supports for each analysis are indicated at the nodes, with NJ on the left and ML on the right.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen YY. 1980. systematic studies on the fishes of the family Homalopteridae of China III. Phyletic studies of the Homalopterid fishes. Zool Systemat. 2:200–211.
- Fang PW. 1935. Study on cross-ostomoid fishes of China. Sinensia. 6:44–97.
- Kottelat M. 2012. Conspectus cobitinum: an inventory of the loaches of the world (Teleostei: Cypriniformes: Cobitoidei). Raffles Bull Zool. 26:1–199.
- Novak J, Hanel L, Rican O. 2006. Formosania: a replacement name for Crossostoma Sauvage, 1878 (Teleostei), a junior homonym of Crossostoma Morris & Lycett, 1851 (Gastropoda). Cybium. 30:92–92.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world. 5th ed. New York (NY): John Wiley and Sons, Inc.
- Tang WQ, Chen YY. 2000. Study on taxonomy of Homalopteridae. J Shanghai Ocean Univ. 9:1–10.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zheng CY. 1991. A taxonomic study on the genus Crossostoma (Pisces:Homalopteridae) with description of a new species. J Jinan Univ (Natural SciMed). 01:77–82.
