Abstract
In this study, we first sequenced and described the complete mitochondrial genome of Pomatorhinus gravivox. The whole genome of P. gravivox was 16,963 bp in length and contained 13 protein-coding genes, 22 transfer RNA genes, two ribosome RNA genes, and one non-coding control region. The overall base composition of the mitochondrial DNA was 29.10% for A, 23.12% for T, 14.56% for G, and 33.22% for C, with a GC content of 47.78%. A phylogenetic tree confirmed that P. gravivox was sister to P. ruficollis and belonged to family Timaliidae. This information will be useful in the current understanding of the phylogeny and evolution of Timaliidae.
The Black-streaked Scimitar Babbler (Pomatorhinus gravivox) is resident bird distributed in China and Southeast Asia, commonly inhabits in shrubs, thorn bushes, and forest margins. This species was split from P. erythrocnemis (Collar Citation2006), and presently five subspecies are recognized as P. g. gravivox, P. g. cowensae, P. g. dedekensi, P. g. decarlei, and P. g. odicus respectively (BirdLife International Citation2018). Pomatorhinus gravivox is listed as of Least Concern (LC) on the IUCN Red List of Threatened Species, and its population is increasing (IUCN, 2018). However, very few genetic studies of P. gravivox had been reported so far. Therefore, we sequenced the complete mitochondrial genome of P. gravivox used next-generation sequencing to enhance our understanding of the phylogeny and evolution of this species in the family Timaliidae.
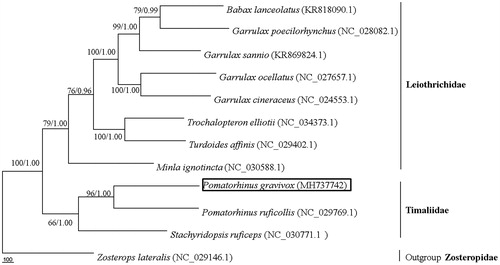
The specimen was collected from Ya’an that located at central region of Sichuan Province in China and stored at the Key Laboratory for Conserving Wildlife with Small Populations in Yunnan, Southwest Forestry University. The total mitochondrial DNA was extracted from the muscle tissue and sequenced using the Illumina Miseq Platform (Illumina, San Diego, CA). The complete mitochondrial genome of P. gravivox was submitted to the NCBI database under the accession number MH737742. Phylogenetic tree of the relationships among Timaliidae and Leiothrichidae were presented using 12 species by maximum parsimony analyses using PAUP* version 4.0b10 software with 1000 bootstrap replication (Swofford Citation2002). Bayesian inference was calculated with MrBayes3.1.2 with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck Citation2003). Sequences of Family Zosteropidae obtained from GenBank (NC030771) were used as outgroup to root trees following Moyle et al. (Citation2012) and Cai et al. (Citation2019).
The complete mitochondrial genome of P. gravivox was 16,963 bp in length. A total of 38 mitochondrial genes were identified, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one non-coding control region (D-loop). Among these genes, ND6 and 8 tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu) were located on the light strand (L-strand), whereas all of the remaining genes were located on the heavy strand (H-strand). The overall base composition of P. gravivox mitogenome was 29.10% for A, 23.12% for T, 14.56% for G, and 33.22% for C. A + T content is 52.22%, which is higher than G + C content of 47.78%, similar to that of the other species in Passeriformes (Huang et al. Citation2016; Li et al. Citation2016; Qi et al. Citation2016; Zhou et al. Citation2016).
The reconstructed phylogenetic tree supported the placement of P. gravivox in family Timaliidae (). All of the nodes were inferred with strong support by the maximum parsimony and Bayesian posterior probabilities analysis. Our results confirmed that P. gravivox located in family Timaliidae was sister to P. ruficollis, which was consistent with BI analysis results of Collar (Citation2006), similar to Moyle et al. (Citation2012) and Cai et al. (Citation2019). In all, the mitochondrial genome reported here would be useful in the current understanding of the phylogeny and evolution of Timaliidae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Birdlife International. 2018. Species factsheet: Pomatorhinus gravivox. [Downloaded on 2018 Nov 30]. http://www.birdlife.org
- Cai TL, Cibois A, Alstrom P, Moyle RG, Kennedy JD, Shao S, Zhang R, Irestedt M, Ericson P, Gelang M, et al. 2019. Near-complete phylogeny and taxonomic revision of the world’s babblers (Aves: Passeriformes). Mol Phylogenet Evol. 130:346–356.
- Collar NJ. 2006. A partial revision of the Asian babblers (Timaliidae). Forktail. 22:85–112.
- Huang R, Zhou YY, Yao YF, Zhao B, Zhang YL, Xu HL. 2016. Complete mitochondrial genome and phylogenetic relationship analysis of Garrulax affinis (Passeriformes, Timaliidae). Mitochondrial DNA A. 27:3502–3503.
- IUCN. 2018. The IUCN Red List of Threatened Species. Version 2018-1. [accessed 2018 Nov 20]. www.incnredlist.org
- Li B, Yao YF, Li DY, Ni QY, Zhang MW, Xie M, Xu HL. 2016. Complete mitochondrial genome of Minla ignotincta (Passeriformes: Timaliidae). Mitochondrial DNA Part B. 1:140–141.
- Moyle RG, Andersen MJ, Oliveros CH, Steinheimer FD, Reddy S. 2012. Phylogeny and biogeography of the core babblers (Aves: Timaliidae). Syst Biol. 61:631–651.
- Qi Y, Zhou YY, Yao YF, Huan ZJ, Li DY, Xie M, Ni QY, Zhang MW, Xu HL. 2016. The complete mitochondrial genome of Babax lanceolatus (Passeriformes: Timaliidae). Mitochondrial DNA A. 27:2925–2926.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Massachusetts.
- Zhou YY, Wei DJ, Qi Y, Xu HL, Li DY, Ni QY, Zhang MW, Yao YF. 2016. Complete mitochondrial genome of Garrulax elliotii (Passeriformes, Timaliidae). Mitochondrial DNA A. 27:3687–3688.