Abstract
The complete mitochondrial genome of Stichaeus grigorjewi (Zoarcales: Stichaeidae) was sequenced for the first time based on the 27 pairs of newly designed primers using Sanger dideoxy sequencing method. Genome structure is consistent with standard scheme known for the vertebrates including 13 gene fragments, two rRNAs, 22 tRNAs, and control region. Moreover, it has additional tRNAs with atypical codons. These features were previously found in Xiphister atropurpureus, which shows the closest position to S. grigorjewi as seen from the phylogram built on the sequences of complete mitochondrial genome.
Keywords:
Zoarcales is a perciform infraorder (Teleostei) comprising about 405 species of coldwater marine fishes (Nelson et al. Citation2016) whose rank and taxonomic composition could be reevaluated in the light of recent molecular phylogenetic findings (Miya et al. Citation2003; Near et al. Citation2012; Radchenko Citation2015; Radchenko Citation2016; Turanov et al. Citation2017). Currently, a less is known about mitochondrial genomics of zoarcoids. However, recently based on the available sequences of complete mitochondrial genome (e.g., Johnstone et al. Citation2007; Swanburg et al. Citation2016; Yang et al. Citation2016; Ayala et al. Citation2017) obvious constrains were found on the nucleotide variation in the control region versus protein-coding genes (Turanov et al. Citation2018). In addition, mitochondrial genomics approaches have provided the opportunity to resolve phylogeographic issues of some endangered zoarcoid species (Lait and Carr Citation2018), demonstrating the possibility of forthcoming extension of its applications for these and other fish taxa. In the current short note, we describe the first complete mitochondrial genome of S. grigorjewi, the prickleback fish (Zoarcales: Stichaeidae) distributed along the coast of the Sea of Japan, Yellow Sea, and the Sea of Okhotsk (Parin et al. Citation2014; Fricke et al. Citation2018).
Total genomic DNA was extracted from the white muscle tissue of the female specimen (MIMB 36611) collected in Vostok Bay of the Sea of Japan (42.89° N, 132.73° E) by gill nets in April 26, 2017. The mitochondrial genome was obtained using Sanger dideoxy sequencing method based on the set of newly designed universal primers by which 27 overlapping regions were amplified (sequences are available upon request). Primer design was performed using Sliding Window-Based PSO Algorithm (Yang et al. Citation2011) based on the sequences available in GenBank for zoarcoids. This approach was recently applied to obtain mitochondrial genome sequences for some fishes (Balakirev, Kravchenko, et al. Citation2018; Balakirev, Romanov, et al. Citation2018).
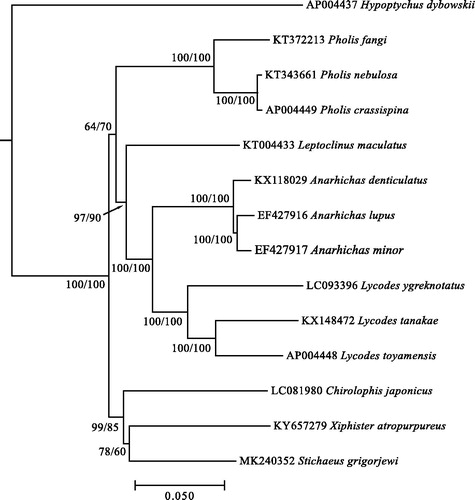
The resulted chromatograms were assembled into mitochondrial genome fragments using Geneious Trial (Kearse et al. Citation2012). Then, the produced consensus sequence of complete genome was annotated using MitoAnnotator online pipeline (Iwasaki et al. Citation2013) and MITOS Web Server (Bernt et al. Citation2013). To clarify the relative position of S. grigorjewi within zoarcoids, we reconstructed NJ-phylogenetic tree using APE R package (Paradis et al. Citation2004). The tree was built based on the sequence matrix consisted of 13 zoarcoid species together with Hypoptychus dybowskii as outgroup with prior alignment in MEGA 6.0 (Tamura et al. Citation2013) using Muscle algorithm (Edgar Citation2004). TrN + G+I nucleotide substitution model implemented in the analysis was found through “modeltest” function of APE.
The mitochondrial genome of S. grigorjewi (GenBank accession number MK240352) is 16,531 bp long, with following overall nucleotide base composition: T (27.5%), C (28.1%), A (26.6%), and G (17.8%). Genome annotation resulted in 13 protein genes where ND6 (and eight tRNAs) reside on the light-strand. All other genes encoding on the heavy strand. It contains 22 tRNAs together with additional tRNA-Leu and tRNA-Ser fragments both maintaining atypical codons as it was also revealed in Xiphister atropurpureus (Ayala et al. Citation2017). There were found two typical rRNAs (16S rRNA and 12S rRNA) and control region, 862 pb in length.
Molecular phylogenetic reconstruction () shows that S. grigorjewi is most closely related to X. atropurpureus and C. japonicus comprising distinct clade within zoarcoids.
Disclosure statement
The authors have no conflicts of interest to declare. The authors are solely responsible for the content and the writing of this paper.
Additional information
Funding
References
- Balakirev ES, Kravchenko AY, Romanov NS, Ayala FJ. 2018. Complete mitochondrial genome of the Arctic rainbow smelt Osmerus dentex (Osmeriformes, Osmeridae). Mitochondrial DNA B. 3:881–882.
- Balakirev ES, Romanov NS, Ayala FJ. 2018. Complete mitochondrial genome of the surf smelt Hypomesus japonicus (Osmeriformes, Osmeridae). Mitochondrial DNA B. 3:1071–1072.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Fricke R, Eschmeyer WN, van der Laan R. 2018. Catalog of fishes: genera, species, references. Institute for Biodiversity Science and Sustainability, California Academy of Sciences. [accessed 2018 Nov 1].
- Ayala L, Becerra J, Boo GH, Calderon D, DiMarco A, Garcia A, Gonzales R, Hughey JR, Jimenez GA. 2017. The complete mitochondrial genome of Xiphister atropurpureus (Perciformes: Stichaeidae). Mitochondrial DNA B. 2:161–162.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Johnstone KA, Marshall HD, Carr SM. 2007. Biodiversity genomics for species at risk: patterns of DNA sequence variation within and among complete mitochondrial genomes of three species of wolffish (Anarhichas spp.). Can J Zool. 85:151–158.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lait LA, Carr SM. 2018. Intraspecific mitogenomics of three marine species-at-risk: Atlantic, spotted, and northern wolffish (Anarhichas spp.). Genome [Internet]. 61:625–634. Available from: https://doi.org/10.1139/gen-2018-0043
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138.
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA. 109:13698–13703.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world. [Place unknown]: John Wiley & Sons.
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 20:289–290.
- Parin NV, Evseenko SA, Vasil’eva ED. 2014. Fishes of the Russian Seas: annotated catalogue. Moscow: KMK Scientific Press.
- Radchenko OA. 2015. The system of the suborder Zoarcoidei (Pisces, Perciformes) as inferred from molecular genetic data. Russ J Genet. 51:1096–1112.
- Radchenko OA. 2016. Timeline of the evolution of eelpouts from the suborder Zoarcoidei (Perciformes) based on DNA variability. J Ichthyol. 56:556–568.
- Swanburg T, Horne JB, Baillie S, King SD, McBride MC, Mackley MP, Paterson IG, Bradbury IR, Bentzen P. 2016. Complete mitochondrial genomes for Icelus spatula, Aspidophoroides olrikii and Leptoclinus maculatus: pan-Arctic marine fishes from Canadian waters. Mitochondrial DNA A. 27:2982–2983.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Turanov SV, Lee Y-H, Kartavtsev YP. 2018. Structure, evolution and phylogenetic informativeness of eelpouts (Cottoidei: Zoarcales) mitochondrial control region sequences. Mitochondrial DNA A [Internet]. 1–9. Available from: https://doi.org/10.1080/24701394.2018.1484117
- Turanov SV, Kartavtsev YP, Lee YH, Jeong D. 2017. Molecular phylogenetic reconstruction and taxonomic investigation of eelpouts (Cottoidei: Zoarcales) based on Co-1 and Cyt-b mitochondrial genes. Mitochondrial DNA A DNA Mapping Seq Anal. 28:547–557.
- Yang C-H, Chang H-W, Ho C-H, Chou Y-C, Chuang L-Y. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729.
- Yang H, Bao X, Wang B, Liu W. 2016. The complete mitochondrial genome of Chirolophis japonicus (Perciformes: Stichaeidae). Mitochondrial DNA A. 27:4419–4420.