Abstract
Landes goose (LG) is a famous fatty liver breed in the world. It is the first time that the complete mitochondrial genome sequence of the LG was reported. The total length of the mtDNA is 16,743 bp, it contains the typical structure, including 22 transfer RNA genes, two ribosomal RNA genes, 13 protein-coding genes, and one non-coding control region (D-loop region). The overall composition of the mtDNA was estimated to be 30.10% for A, 22.57% for T, 32.19% for C, and 15.14% for G. Phylogenetic analyses using N-J computational algorithms showed that the analyzed 18 Anseriform species are divided into four major clades: Anatinae, Anserinae, Dendrocygninae, and Anseranatidae. In addition, our work confirmed that LG and Anser anser have a close genetic relationship with fellow tribal members Anser fabalis.
Landes goose (LG) is a famous fatty liver breed in the world (Tang et al. Citation2018). In this study, we newly determined the complete mitochondrial genome of LG, and the adult individuals of LG were collected at its culturing farm in Yuanjiang city (28.50 N and 112.20 E), Hunan province, China on September 2018. And, the specimens were stored at −80 °C in our laboratory (Research Department of Animal Nutrition and Poultry, Hunan Institute of Animal and Veterinary Science). Total genomic DNA was extracted from the thigh muscle of a single individual using the EasyPure Kit of Genomic DNA (Transgen Biotech, Beijing, China). Whole mitochondrial genome was amplified with 13 pairs of primers and sequenced using BioSune Biotech (Shanghai, China). DNA sequence was analyzed using DNAStar.Lasergene.v7.1 software (Madison, WI), tRNA Scan-SE1.21 software (Lowe and Eddy Citation1997), and DOGMA software (Wyman et al. Citation2004).
The total length of the mtDNA is 16,743 bp, it contains the typical structure, including 22 transfer RNA genes, two ribosomal RNA genes, 13 protein-coding genes, and one non-coding control region (D-loop region) (GenBank accession No. MK133021). The overall composition of the mtDNA was estimated to be 30.10% for A, 22.57% for T, 32.19% for C, and 15.14% for G, and the percentage of A and T (52.67%) was higher than that of G and C (47.33%). All the protein initiation codons are ATG, except for COX1, COX2, and ND5 are GTG. All these genes have 16 spaces and eight overlaps both in the length of 1–10 bp. These genes had five types of termination codons, including TAA, TAG, AGG, AGA, and an incomplete termination codon ‘‘T– –’’. ‘‘T– –’’ is the 5' terminal of the adjacent gene (Anderson et al. Citation1981). Among 13 protein-coding genes, the longest one was ND5 gene (1818 bp), which was located between the tRNALeu and Cytb, and the shortest one was ATPase8 gene (168 bp), which was located between the tRNALys and ATPase6. The lengths of 12S rRNA and the 16S rRNA were 988 bp and 1610 bp. And, deduced 22 tRNA genes were distributed in rRNA and protein-coding genes, ranging from 66 to 74 bp in size. The mitochondrial DNA D-loop region of the LG was located between tRNAGlu and tRNAPhe with a length of 1179 bp.
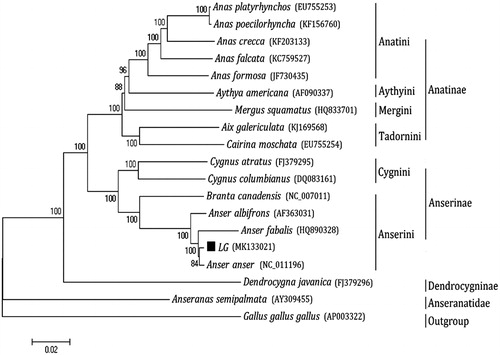
Phylogenetic analysis was performed using the complete mitochondrial DNA sequences of 18 Anseriformes. Each of the sequence dataset was aligned using ClustalX (Thompson et al. Citation1997) and analyzed by neighbor-joining (N-J) in MEGA 4.0 (Tamura et al. Citation2007), and bootstrap analysis was performed with 100 replications. An N-J tree showed that the analyzed species are divided into four major clades (). Anatinae makes up the first lineage, which is sister to the second group, Anserinae; Dendrocygninae forms the third group and is sister to Anserinae and Anatinae. The lineage consisting of these three groups, in turn, is sister to the fourth clade, Anseranatidae. In addition, our work confirmed that LG and Anser anser have a close genetic relationship with fellow tribal members Anser fabalis.
Figure 1. Phylogenetic analysis based on complete mitochondrial genome sequences. An N-J tree was built based on the phylogenetic analysis of 18 Anseriform species’ complete mitochondrial genomes. The mitochondrial genome sequences of the Anseriform species were obtained from the GenBank databases (Accession numbers have marked on the figure). Abbreviation of species indicates LG, Landes goose.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–464.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 24:1596–1599.
- Tang J, Fang Q, Shao R, Shen J, He J, Niu D, Lu L. 2018. Digital gene-expression profiling analysis of the fatty liver of Landes geese fed different supplemental oils. Gene. 673:32–45.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
