Abstract
In this study, we first determined and described the mitochondrial genome of Tridacna crocea, a member of the subfamily Tridacninae. The existing mitogenome of T. crocea is 19,157 bp in length, containing 13 protein-coding genes, 2 rRNA genes, 24 tRNA genes, and 1 non-coding control region. Phylogenetic analysis revealed that T. crocea was closely related to T. squamosa.
Tridacna crocea (Lamarck Citation1819) belongs to Mollusca, Bivalvia, Cardiida, Cardiidae, Tridacninae, which is mainly present in tropical and subtropical shallow sea coral reef environments of Indo-West Pacifica region (Neo et al. Citation2015). This species has been listed in the Convention on International Trade in Endangered Species (CITES) appendix II (https://cites.org/eng), and International Union for Conservation of Nature (IUCN) Red List of Threatened Species (Version 2017-2, www.iucnredlist.org). Here, we first sequenced and characterized the mitochondrial genome of T. crocea, and examined the phylogenetic position of T. crocea within Tridacninae.
The specimen of T. crocea was collected from Niuche bay, Sanya, Hainan Province, China. Only small pieces of mantle tissues were collected, which is not lethal for the giant clams (Kochzius and Nuryanto Citation2008). The total genomic DNA was extracted and stored at −80 °C in Institute of Deep-sea Science and Engineering, CAS. The mitochondrial genome of T. crocea was sequenced using 6 pairs of degenerate primer. The annotation of mitochondrial protein-coding genes and ribosomal RNA genes were identified using online service ORF-Finder (https://www.ncbi.nlm.nih.gov/orffinder/) (Rombel et al. Citation2002) and MITOS webserver (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). tRNAscan-SE (Lowe and Eddy Citation1997) was used to predict the transfer RNA genes. The T. crocea mitogenome has been deposited in the GenBank database (accession number: MK249738).
The mitochondrial genome of T. crocea is 19,157 bp in length, containing 13 protein-coding genes, 2 rRNA genes, 24 tRNA genes, and 1 non-coding control region. Because there are many repeated sequences, only partial sequences are obtained in the non-coding region. The overall basic composition of the heavy strand in T. crocea is 26.1% A, 35.8% T, 23.1% G and 15.0% C, with an AT content of 61.9%. All protein-coding genes start with a typical ATN codon, except for cytb which started with TTG. Eleven protein-coding genes stop with TAN, while cytb and nad2 stop with TGG and TTA, respectively.
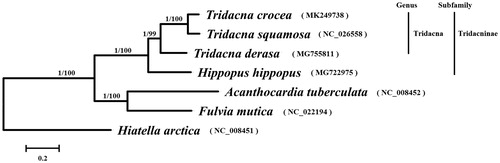
Seven shellfish mitogenomes including the one obtained in this study were used for phylogenetic analysis. Hiatella arctica was used as an outgroup. Phylogenetic reconstruction was performed using Bayesian Inference (BI) and maximum likelihood (ML) with MrBayes v3.2 (Ronquist et al. Citation2012) and MEGA v6.0 (Tamura et al. Citation2013), respectively. Both BI and ML analyses produced identical tree topologies with varying levels of support (). Phylogenetic analysis revealed that T. crocea was closely related to T. squamosa. This study will provide valuable molecular data for further evolutionary analysis of this threatened species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Kochzius M, Nuryanto A. 2008. Strong genetic population structure in the boring giant clam, Tridacna crocea, across the Indo-Malay Archipelago: implications related to evolutionary processes and connectivity. Mol Ecol. 17:3775–3787.
- Lamarck J-B. M d. 1819. Histoire naturelle des animaux sans vertèbres. Tome sixième, 1re partie. Paris: Verdière, vi + 343 pp.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Neo ML, Eckman W, Vicentuan K, Teo SLM, Todd PA. 2015. The ecological significance of giant clams in coral reef ecosystems. Biol Conserv. 181:111–123.
- Rombel IT, Sykes KF, Rayner S, Johnston SA. 2002. ORF-FINDER: a vector for high-throughput gene identification. Gene. 282:33–41.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice ccross a large model space. Syst Biol. 61:539–542.
- Tamura K, Stecher G, Peterson D, Filipski A, kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.