Abstract
The complete mitochondrial genome of the Yunnanilus sichuanensis (Cypriniformes: Nemacheilinae) was first sequenced using the next-generation sequencing in this study. The mitogenome was 16,571 bp in length and contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, 1 displacement loop (D-loop) locus, and an origin of replication on the light-strand (OL) region. Most of the genes were encoded on the heavy strand except ND6 and 8 tRNA genes encoded on the light strand. The mitogenome organization and gene order were similar to a typical vertebrate mitogeneome. The overall base composition was 26.96% A, 28.62% T, 25.75% C and 18.67% G, with 55.58% AT. Phylogenetic analysis based on the 13 protein-coding genes nucleotide sequences with two different methods (neighbour-joining and maximum likelihood analysis) showed that Y. sichuanensis was nearest to Y. jinxiensis, coming together into a branch, meanwhile together with Triplophysa venusta formed a sister group. This study enriched the data of the mitogenome of the genus Yunnanilus, and provide an important information for morphological classification of Nemacheilinae species and the study of biological origin.
Yunnanilus sichuanensis (Cypriniformes: Nemacheilinae) is endemic to the Upper Yangtze River in China, which is mainly distributed in the slow water area of river trunk and branch flow, with an altitude of l500–2300 mm (Ding Citation1995). Currently, only the mitogenome of Y. jinxiensis (GenBank Accession number MG 818720) in the same genus has been reported. Recent research has indicated that the natural population number of this fish strongly decreased due to the construction of hydroelectric dams, habitat loss, overfishing and environmental pollution (Ding Citation1995).
The complete mitogenome of Y. sichuanensis was first sequenced using the next-generation sequencing (NGS) in this study. The specimens were collected from Guanghan, Sichuan province of China (30°52′51.41″N, 104°14′55.77″E) in September 2018 and were stored in Zoological Specimen Museum of Neijiang Normal University (accession number: 20180925BB102). A 30–40 mg fin clip was collected and preserved in 95% ethanol at 4 °C. Total genomic DNA was extracted from these caudal fins by a Tissue DNA Kit (OMEGA E.Z.N.A., Norcross, GA, USA) following the manufacturer’s protocol. Subsequently, the genomic DNA was sequenced using the NGS, and then the mitogenome was assembled using Y. jinxiensis as a reference.
The complete mitogenome of Y. sichuanensis was a circular molecule of 16,571 bp in length (GenBank Accession number MK181571), including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, 1 displacement loop (D-loop) locus, and an origin of replication on the light-strand (OL) region, which was in conformity with typical vertebrate mitogeneome (Sun et al. Citation2014; Murienne et al. Citation2016). Most of the genes were encoded on the heavy strand except ND6 and 8 tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro) encoded on the light strand. The overall base composition was 26.96% A, 28.62% T, 25.75% C, and 18.67% G, with a slight A + T bias of 55.58%. Thirteen intergenic spacers (total 68 bp) varying from 1 to 31 bp and 9 gene overlaps (total 30 bp) ranging from 1 to 10 bp existed in the mitogenome. The 13 protein-coding genes vary from 1839 bp (ND5) to 168 bp (ATP8) in length. The common initiation codon was ATG in the protein-coding genes, except for COI, which started with GTG. TAA and TAG were usual two stop codons in 10 genes (TAA: ND1, COI, ATP8, ATP6, ND4L; TAG: ND2, ND3, ND4, ND5, ND6). The rest of genes ends with an incomplete termination codon T (COII, COIII, Cyt b). In addition, all the tRNA genes length ranging from 66 to 76 bp, the tRNACys was the shortest while tRNALys was the longest. The 12S rRNA gene (950 bp) and 16S rRNA gene (1677 bp) were located between the tRNAPhe and tRNALeu genes, separated by the tRNAVal gene. The two main non-coding distinctions in the Y. sichuanensis mitogenome were D-loop (916 bp) and OL (31 bp). Furthermore, D-loop was located between tRNAPro and tRNAPhe, while OL was found in between tRNAAsn and tRNACys.
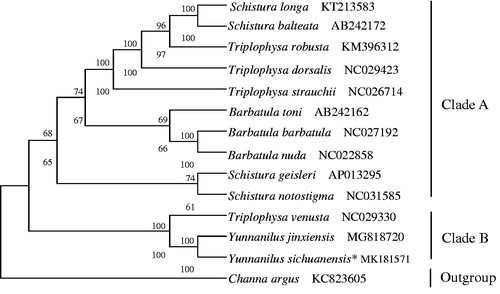
Phylogenetic tree has been known as a model of biological evolution, and its establishment is almost all based on amino acid sequences or DAN sequences (Chor and Tuller Citation2005). Neighbor-joining (NJ) and maximum likelihood (ML) methods are two common methods to construct the phylogenetic tree (Dong et al. Citation2008; Zou et al. Citation2018). Additionally, ML is an increasingly popular optimality criterion for selecting evolutionary trees (Chor and Tuller Citation2005). To confirm the phylogenetic relationships between Y. sichuanensis and other Cobitidae family fishes, phylogenetic analysis were performed on the concatenated dataset of 13 protein-coding genes (PCGs) at nucleotide level with NJ and ML (Li et al. Citation2016; Zou et al. Citation2017). There were 14 species in the phylogenetic tree, including Channa argus (Channoidei, Channidae) as an outgroup. The tree topologies produced by NJ had nearly the same topology as that of ML tree (). Both trees showed that Y. sichuanensis was nearest to Y. jinxiensis, coming together into a branch, meanwhile together with Triplophysa venusta formed Clade B, and the rest of species were clustered into clade A. Additionally, results suggested that Yunnanilus has a closer relationship with Triplophysa than Schistura and Barbatula . These results may provide an important information for morphological classification of Nemacheilinae species and the study of biological origin.
Figure 1. The phylogenetic analyses investigated using neighbor-joining (NJ) and maximum likelihood (ML) methods analysis indicated evolutionary relationships among 14 taxa based on the nucleotide sequences of 13 concatenated protein-coding genes. The yielded NJ tree had a same topology as that of ML tree. NJ posterior probability and ML bootstrap support values are shown on the nodes. Channa argus (GenBank: KC823605) was used as an outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chor B, Tuller T. 2005. Maximum likelihood of evolutionary trees: hardness and approximation. Bioinformatics. 21:i97–i106.
- Ding RH. 1995. A new species of the genus Yunnanilus from western Sichuan, China (Cypriniformes: Cobitidae). Zool Systemat. 20:253–256.
- Dong AG, Gao L, Zhao JB, Hu JL. 2008. Construction of phylogenetic tree based on DNA sequences. J Northwest A & F Univ. 36:221–226.
- Li S, Guo WT, Liu XQ, Qu HT, Guan M, Su W. 2016. Complete mitochondrial genome of the hybrid of Acipenser schrenckii (♀) × Huso dauricus (♂). Mitochondrial DNA. 27:7–8.
- Murienne J, Jeziorski C, Holota H, Coissac E, Blanchet S, Grenouillet G. 2016. PCR-free shotgun sequencing of the stone loach mitochondrial genome (Barbatula barbatula). Mitochondrial DNA A. 27:4211–4212.
- Sun QL, Wang DQ, Wei QW, Wu JM. 2014. The complete mitochondrial gemone of Triplophysa robusta (Teleostei, Cypriniformes: Cobitidae). Mitochondrial DNA. 27:1.
- Zou YC, Xie BW, Qin CJ, Wang YM, Yuan DY, Li R, Wen ZY. 2017. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 39:767–778.
- Zou YC, Zhang JY, Xie M, Zhang T, Deng Q, Wen ZY. 2018. The complete mitochondrial genome of Ancherythroculter wangi and its phylogeny. Mitochondrial DNA Part B. 3:276–277.
