Abstract
Morella salicifolia (Myricaceae) is a dioecious plant of medicinal value. This study sequenced the complete chloroplast genome sequence of Morella salicifolia, the second to be sequenced in Myricaceae. The genome is 159,010 bp in size, composed of two copies of inverted repeat (IR) regions (26,045bp each) separating a large single-copy (LSC) region (88,207bp) from the small single-copy (SSC) region (18,713bp). The genome has 112 genes (77 protein-coding, 4 rRNA, and 31 tRNA) and an overall GC content of 36.2%. Phylogenetic relationship clustered M. salicifolia with M. rubra and Myricaceae clustered with Juglandaceae.
Introduction
Morella salicifolia (Hochst. ex A.Rich.) Verdc. & Polhill (Myricaceae) is a dioecious plant of medicinal applications, e.g. analgesic (Njung'e et al. Citation2002), treating gastro-intestinal disorders (Kefalew et al. Citation2015), digestive problems (Silva et al. Citation2015), and headache (Teklay et al. Citation2013). Previously, it was under the genus Myrica before the genus being split into two, Myrica and Morella (Herbert Citation2005). These two genera are taxonomically close (Silva et al. Citation2015) and Morella is the largest containing ∼50 species (Herbert Citation2005) distributed in Europe, North America, Africa, and Asia. However, there exist taxonomic challenges in the position of Morella in Myricaceae and the entire family in Fagales (Liu et al. Citation2017). In Myricaeae, only one chloroplast genome (M. rubra: NC_035006) has been reported (Liu et al. Citation2017), despite the chloroplast being proven to be valuable for molecular genetic studies in plants due to its conserved gene order (Deguilloux et al. Citation2004). This study sequenced the chloroplast genome of M. salicifolia and constructed the phylogenetic relationship with other Fagales.
Leaf samples of M. salicifolia were obtained from Mount Rungwe Nature Forest Reserve (Mbeya, Tanzania; co-ordinates: 9.237392°S, 33.98566°E, Voucher Number: HIB-MS118). Genomic DNA was extracted using CTAB procedure, then sequenced using Illumina Hiseq 2500 platform (Novogene Technologies, Inc. Beijing, China). De novo assembly produced a total of 5.2 Gb raw data which were filtered by PRINSEQ lite Ver0.20.4 and the high-quality reads (∼5G) assembled by NOVOPlasty 2.7.0 with K-mer 31–39 using reference chloroplast genome of M. rubra (NC_035006). Gene annotation was done by online Dual Organellar GenoMe Annotator DOGMA (Wyman et al. Citation2004), and manual amendments based on BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Moreover, the tRNA genes were verified using tRNAscan-SE 2.0 (Lowe and Chan Citation2016) and the annotated genome sequence was submitted to NCBI GenBank (Accession number MK310272).
The chloroplast genome of M. salicifolia is 159,010 bp in length, comprising a pair of IRs 26,045 bp in length, separating the LSC 88,207 bp and SSC 18,713 bp regions. Overall, 112 unique genes were annotated including 77 protein-coding genes, 4 rRNA genes, and 31 tRNA genes. Six protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7, rps12), seven tRNA genes (trnI-CAU, trnV-GAC, trnA-UGC, trnL-CAA, trnI-GAU, trnR-ACG & trnN-GUU), and four rRNA genes were duplicated within the IRs. The nucleotide content was 36.2% (GC) and 63.8% (AT). In addition, 17 genes contain introns with 15 having one intron, while 2 had two introns (ycf3 & clpP). The genome was observed to have lost the infA gene and pseudogenization of rpl22 and ycf15 genes.
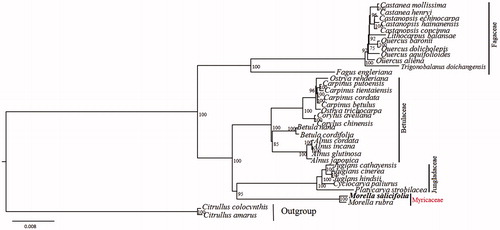
To assess the phylogenetic position of Morella in Fagales, 34 chloroplast genomes were downloaded from the NCBI, including 32 fagales and 2 cucurbitales as outgroup. A Maximum Likelihood (ML) phylogenetic tree was developed in RAxML v8.1.11 (Stamatakis Citation2014) via the best-fit substitution model (GTR + I + G) obtained from jModelTest v2.1.4 (Darriba et al. Citation2012). The tree revealed that M. salicifolia clustered with M. rubra and Myricaceae clustered with Juglandaceae (). This study provides genomic resources for Myricaceae, a gap which has hindered the resolution of its evolution in Fagales.
Figure 1. The Maximum Likelihood (ML) phylogenetic tree of M. salicifolia and related species based on complete chloroplast genomes. Numbers in the nodes represent ML bootstrap support values. Accession numbers: Castanea mollissima NC_014674, C. henryi NC_033881, Castanopsis echinocarpa NC_023801, C. hainanensis NC_037389, C. concinna NC_033409, Lithocarpus balansae NC_026577, Quercus baronii NC_029490, Q. dolicholepis NC_031357, Q. aquifolioides NC_026913, Q. aliena NC_026790, Trigonobalanus doichangensis NC_023959, Fagus engleriana NC_036929, Ostrya rehderiana NC_028349, Carpinus putoensis NC_033503, C. tientaiensis NC_034910, C. cordata NC_036723, C. betulus NC_036723, Ostrya trichocarpa NC_034295, Corylus avellana NC_031855, C. chinensis NC_032351, Betula nana NC_033978, B. cordifolia NC_037473, Alnus cordata NC_036751, A. incana NC_036752, A. glutinosa NC_039930, A. japonica NC_036753, Juglans cathayensis NC_033893, J. cinereal NC_035960, J. hindsii NC_035965, Cyclocarya paliurus NC_034315, Platycarya strobilacea NC_035413, Morella rubra NC_035006, Citrus colocynthis NC_035727, C. amarus NC_035974.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Darriba D, Taboada G, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 9:772.
- Deguilloux M, Pemonge M, Petit R. 2004. Use of chloroplast microsatellites to differentiate oak populations. Ann Forest Sci. 61:825–830.
- Herbert J. (2005). Systematics and biogeography of Myricaceae [Unpublished Dissertation]. St. Andrews (Scotland): University of St. Andrews.
- Kefalew A, Asfaw Z, Kelbessa E. 2015. Ethnobotany of medicinal plants in Ada'a District, East Shewa Zone of Oromia Regional State, Ethiopia. J Ethnobiol Ethnomed. 11:25.
- Liu L, Li R, Worth J, Li X, Li P, Cameron K, Fu C. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Lowe T, Chan P. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Njung'e K, Muriuki G, Mwangi J, Kuria K. 2002. Analgesic and antipyretic effects of Myrica salicifolia (Myricaceae). Phytother Res. 16:73–74.
- Silva B, Seca A, Barreto C, Pinto D. 2015. Recent Breakthroughs in the antioxidant and anti-inflammatory effects of Morella and Myrica species. Int J Mol Sci. 16:17160–17180.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Teklay A, Balcha A, Mirutse G. 2013. An ethnobotanical study of medicinal plants used in Kilte Awulaelo District, Tigray Region of Ethiopia. J Ethnobiol Ethnomed. 9:65.
- Wyman S, Jansen R, Boore J. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
