Abstract
We sequenced 15,803 bp of the leaf-rolling-weevil, Apoderus jekelii (Coleoptera: Attelabidae) mitochondrial genome (mitogenome) that lacked ∼8000 bp of the A + T-rich region for the completion of the genomic sequence. The A. jekelii mitogenome, which includes 1169 bp of A + T-rich region, possesses typical sets of genes [13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes] and gene arrangement of the species was identical to that commonly found in the majority of the insects. The start codon for ND1 was a rare TTG instead of typical ATN. Nearly, all PCGs ended with complete stop codons, TAA or TAG, except for ND5 which ended with an incomplete T. Phylogenetic analyses using the eight concatenated PCG sequences, which are commonly available for the mitogenome sequences of Curculionoidea, revealed Attelabidae as monophyletic, as well as the sister relationship between current A. jekelii and congeneric species A. coryli in Attelabidae, with the highest nodal supports both in Bayesian inference and maximum likelihood methods. In order to gain a more comprehensive picture of the phylogenetic relationships among the lineages of Attelabidae, an extended analysis with more taxonomic sampling will be necessary.
The leaf-rolling-weevil-weevil, Apoderus jekelii Roelofs, 1874 (Coleoptera: Attelabidae), is distributed in Korea, China, East Siberia, and Japan (Park et al. Citation2007). Adults occur twice per year from May to September (Park Citation2005). It is a forest pest feeding on several species of Alnus (A. japonica, A. hirsuta, A. firma, and A. pendula), heartleaf hornbeam (Carpinus cordata), and the Asian-beaked hazel (Corylus sieboldiana) (Park et al. Citation2007). One of the interesting features of the species includes different leaf-cutting patterns in adults such as a straight-cutting, single-cutting, and non-cutting types (Park et al. Citation2007, Citation2012; Park and Park Citation2014).
An A. jekelii adult was collected at Wangpi-gil, Geumgangsong-myeon, Uljin-gun, Gyeongsangbuk-do, Republic of Korea (36°54'33″ N, 129°14'25″ E) on May 2017. This voucher specimen was deposited at the Chonnam National University, Gwangju, Korea, with the accession number CNU7498. DNA was extracted from the hind legs of the A. jekelii individual using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) and three long overlapping fragments (LFs; COI-ND5, ND5-lrRNA, and lrRNA-COI) were amplified. These LFs were subsequently used as templates to amplify 26 × 500–700 bp long short overlapping fragments. All primers were designed from the available mitogenome sequences of Attelabidae (data not shown). The A. jekelii sequence was deposited in GenBank with the accession number MK292540.
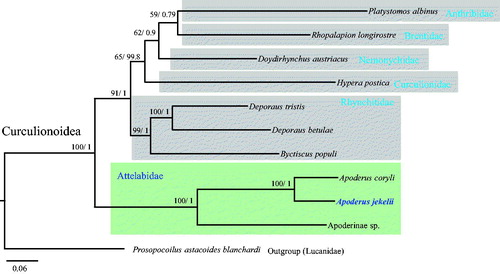
Currently, only two mitogenomes are available for Attelabidae in which A. jekelii is included and even these are incomplete, lacking a substantial portion of the genome (Apoderinae sp. and Apoderus coryli). Thus, a phylogenetic analysis was conducted using the eight commonly available PCGs (5382 bp including gaps) after the well-aligned conserved blocks were selected using Gblocks 0.91b software (Castresana Citation2000), with the inclusion of representative sequences of other phylogenetically close families in Curculionoidea, the sequences of which also lack in some portions of their genomes. Bayesian inference (BI) and maximum likelihood (ML) methods were conducted using MrBayes ver. 3.2.2 (Ronquist et al. Citation2012) and RAxML-HPC2 ver. 8.0.24 (Stamatakis Citation2014), respectively, implemented on the CIPRES Portal ver. 3.1 (Miller et al. Citation2010). Prosopocoilus astacoides blanchardi belonging to Lucanidae in Scarabaeoidea was used as an outgroup. Trees were visualized with FigTree ver. 1.42 (http://tree.bio.ed.ac.uk/software/figtree/) ().
Figure 1. Phylogenetic tree of Curculionoidea. Maximum likelihood (ML) and Bayesian inference (BI) methods produced the same topology based on concatenated eight PCGs (ATP8, ATP6, COIII, ND3, ND5, ND4, ND4L, and ND6). The numbers at each node specify bootstrap percentages from 1000 pseudoreplicates by ML analysis (first value) and Bayesian posterior probabilities in percent from the BI analysis (second value). The scale bar indicates the number of substitutions per site. Lucanidae (Prosopocoilus astacoides blanchardi) was utilized as an outgroup. GenBank accession numbers are as follows: Platystomos albinus, KX087337 (Linard et al. Citation2018); Rhopalapion longirostre, JN163967 (Haran et al. Citation2013); Doydirhynchus austriacus, JN163964 (Haran et al. Citation2013); Hypera postica, JN163953 (Haran et al. Citation2013); Deporaus tristis, KX087280 (Linard et al. Citation2018); Deporaus betulae, JN163945 (Haran et al. Citation2013); Byctiscus populi, JN163965 (Haran et al. Citation2013); Apoderus coryli, JN163966 (Haran et al. Citation2013); Apoderinae sp., MH473531 (Gillett et al. Citation2014), and; Prosopocoilus astacoides blanchardi, KF364622 (Kim et al. Citation2015).
The A. jekelii mitogenome, which includes an unfinished portion of the A + T-rich region, was 15,803 bp and possessed the typical sets of genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes). The arrangement of this genome was identical to that typically observed in other insects (Cameron Citation2014). Previously, the coleopteran mitogenomes with substantially large A + T-rich regions have been sequenced in several species: 5654 bp in Protaetia brevitarsis (Kim et al. Citation2014), 1629 bp in Cicindela anchoralis (Wang et al. Citation2018), 3100 bp in Prosopocoilus astacoides blanchardi (Kim et al. Citation2015), 4469 bp in Coccinella septempunctata (Kim et al. Citation2012), and 1703 bp in Damaster mirabilissimus mirabilissim (Wan et al. Citation2012). At the same time, a substantial number of coleopteran mitogenomes that lack the A + T-rich region and neighboring regions are also registered in the GenBank. This may have occurred because of the difficulty in sequencing the repeat sequences with a high A/T composition as we have also experienced. Twelve PCGs had the typical ATN start codon, whereas ND1 had an atypical TTG codon. Nearly, all PCGs ended with complete stop codons such as TAA or TAG, but ND5 ended with an incomplete T.
Phylogenetic analyses based on the concatenated sequences of eight PCGs placed A. jekelii and congeneric species A. coryli in a sister group with both BI and ML analyses showing the highest nodal support. Three available species of Attelabidae, within which current A. jekelii belongs, formed a strong monophyletic group also with the highest nodal support in both analyses (). Currently, mitogenome sequences are available only from three species including current A. jekelii and the two previously reported species have unfinished genes and regions. Therefore, we expect that our mitogenome sequences of A. jekelii will be used for primer design and further improved annotation of taxonomically close groups, along with a phylogenetic analysis of the Attelabidae, despite our sequence largely missing the A + T-rich region.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Gillett CP, Crampton-Platt A, Timmermans MJ, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31:2223–2237.
- Haran J, Timmermans MJ, Vogler AP. 2013. Mitogenome sequences stabilize the phylogenetics of weevils (Curculionoidea) and establish the monophyly of larval ectophagy. Mol Phylogenet Evol. 67:156–166.
- Kim MJ, Im HH, Lee KY, Han YS, Kim I. 2014. Complete mitochondrial genome of the whiter-spotted flower chafer, Protaetia brevitarsis (Coleoptera: Scarabaeidae). Mitochondrial DNA. 25:177–178.
- Kim MJ, Kim K-G, Kim SR, Kim I. 2015. Complete mitochondrial genome of the two-spotted stag beetle, Metopodontus blanchardi (Coleoptera: Lucanidae). Mitochondr DNA. 26:307–309.
- Kim MJ, Wan X, Kim I. 2012. Complete mitochondrial genome of the seven-spotted lady beetle, Coccinella septempunctata (Coleoptera: Coccinellidae). Mitochondr DNA. 23:179–181.
- Linard B, Crampton-Platt A, Moriniere J, Timmermans MJTN, Andújar C, Arribas P, Miller KE, Lipecki J, Favreau E, Hunter A, et al. 2018. The contribution of mitochondrial metagenomics to large-scale data mining and phylogenetic analysis of Coleoptera. Mol Phylogenet Evol. 128:1–11.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), IEEE, 14 November 2010, New Orleans, LA; p. 1–8.
- Park JY. 2005. Study on systematics and ecology of the Attelabidae (Coleoptera) from Korea [dissertation]. Andong (Gyeongsang Province): Andong National University.
- Park JY, Kwon OS, Lee JE. 2007. Descriptions of larvae and pupae of the subfamily Apoderinae (Coleoptera: Attelabidae) in Korea. Entomol Science. 10:87–102.
- Park JY, Lee JE, Park JK. 2012. Leaf cutting-patterns and general cradle formation process of thirteen Apoderinae (Coleoptera, Attelabidae) in Korea: cradles of Attelabidae in Korea I. Entomol Res. 42:63–71.
- Park JY, Park JK. 2014. Analysis of host preference of subfamily Apoderinae (Coleoptera: Attelabidae) in Korea. Korean J Nat Conserv. 8:39–46.
- Ronquist F, Teslenko M, Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mr. Bayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wan X, Hong MY, Liao A, Kim MI, Kim K-G, Han YS, Kim I. 2012. Complete mitochondrial genome of a carabid beetle, Damaster mirabilissimus mirabilissim (Coleoptera: Carabidae). Entomol Res. 42:44–54.
- Wang AR, Kim MJ, Jeong SY, Kim I. 2018. Complete mitochondrial genome sequence of Cicindela anchoralis Chevrolat, 1845 (Coleoptera: Carabidae). Mitochondr DNA B. 3:282–283.
